OPTICS FOR MICROSCOPY: Swept field confocal overcomes point-scanning microscopy limitations
BILL VOGT, LONG YAN, MICHAEL SZULCZEWSKI, and KEVIN ELICEIRI
Significant advances in our understanding of human health and disease have relied on our ability to identify the location of individual molecules and to study their dynamics within living cells taken from human and animal models. With enhanced optical resolution and optical sectioning capability, confocal laser scanning microscopy (CLSM) has become an essential biological imaging tool. With CLSM the cell biologist can, for instance, examine sub-cellular structures, and the neuroscientist can explore the connectivity among the neuron networks. However, despite its success, the conventional CLSM does have certain limitations in capturing dynamic events such as slow acquisition speed and relatively high phototoxicity.
To record fast biological events, alternative confocal technologies have been developed using a variety of approaches, including a disk with pinholes that splits the excitation beam into multiple beams that can spin over a sample, or a disk with a slit that can sweep light across the sample in the same way a paintbrush sweeps paint across a surface. These alternate confocal approaches greatly improve the imaging speed compared to a traditional confocal point scanner. However, there are some tradeoffs for this speed improvement such as high crosstalk, low spatial resolution, limited scan modes, and limited objective lens selection. Fortunately, a new technology, the swept field confocal (SFC), uses an innovative design to overcome limitations of traditional point-scanning confocal systems and address many of the noted shortcomings of other fast confocal technologies.
Biological relevance
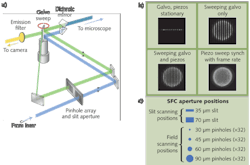
The SFC microscope incorporates both high-resolution pinhole imaging along with high-speed slit imaging modes in a single unit. By using a positionable aperture plate containing a variety of pinhole columns and slit apertures, the excitation light can be split into multiple small beamlets or formed into a ribbon of light as required by each mode. Unlike spinning disk confocal systems that are limited by having their pinhole apertures embedded in a spinning disk, the SFC's aperture plate remains stationary. Galvonometric and piezo-controlled mirrors sweep the image of the selected 32 pinhole column or single slit across the sample. The emission photons are de-scanned and focused through a complementary aperture column or slit and onto a charge-coupled device (CCD) camera. This results in a high-resolution image that can be collected at up to 100 frames per second (fps) in the pinhole mode and greater than 1000 fps in the slit mode.
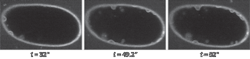
The ability for the scientist to match the optical recording with the temporal biological fluorescence response has great promise for cell and developmental biology studies in which live dynamic events must be captured quickly in high resolution. The SFC has been used in many types of biological studies such as trafficking studies (see Fig. 1). In addition, many other studies have been published with the SFC ranging from protein–protein interactions, mechanotransducer channels, and signaling pathways to exocytosis.1, 2, 3, 4
Scanning field confocal benefits
Key advantages of the SFC design enabled by the imaging optics include high-speed imaging capability, high quantum efficiency (QE) detection and low phototoxicity, variable pinhole sizes or slits, and reduced crosstalk between confocal apertures (see table).Improving the aperture plate
In early designs of the SFC we used a two-dimensional (2D) array with 84 pinhole aperture. Even though we had success in imaging with this 2D pinhole array, there was high crosstalk among pinholes when thicker samples were imaged. This problem is also seen in very successful optical microscopes such as the Yokogawa (Tokyo, Japan) spinning disc confocal.
To overcome this crosstalk issue, we spaced each pinhole and formed a linear array. The linear array has significant advantages over a 2D array: 1) The crosstalk between pinholes is minimized, which means the SFC can be used in thicker samples such as brain slices; and 2) any excitation and emission aperture combination can be designed–we added excitation and emission slits for band scanning as well as high-speed imaging, for example. Dr. Jeff Magee at the Janelia Farm Research Campus (Ashburn, VA), part of the Howard Hughes Medical Institute (www.hhmi.org), obtained remarkable successes with the design we provided.5 Using the linear pinhole array, he was able to image at up to 200 fps in brain slices and with the slits he was able to image at 3000 fps.
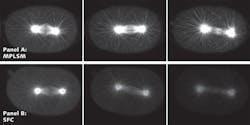
The unique combination of the linear pinhole array to allow slice imaging, together with the ability to band scan and to use high QE detection (CCD cameras), means that high-speed images in thicker samples can be obtained with minimal phototoxicity. The Laboratory for Optical and Computational Instrumentation (LOCI) group at the University of Wisconsin at Madison demonstrated that SFC can be used for trafficking studies where speed, high resolution, and viability are all needed. The SFC method compares favorably to multiphoton microscopy, which had been the gold standard for much of this work (see Fig. 3).Scanning speeds and limitations
The SFC uses an innovative design to achieve real-time video rate speeds during acquisition. The imaging speed of the SFC is largely limited by the CCD camera used. In slit mode the SFC can reach speeds of 700 fps with the Cascade II from Photometrics (Tucson, AZ) and up to 3 kHz when the NeuroCCD-SMQ from RedShirtImaging (Decatur, GA) is used. However, the NeuroCCD-SMQ has poor spatial resolution (80 × 80 pixels) compared to the Cascade II (512 × 512 pixels). In order to achieve high speed without losing spatial resolution, the CCD camera speed needs to be improved, which is an area of active commercial development.
Future directions
With the high speed and high QE of the SFC, there is great interest in further live-cell imaging applications in cell biology and neurosciences. A particular area of interest is to add quantitative capabilities such as fluorescence resonance energy transfer (FRET) and fluorescence lifetime imaging (FLIM) so the SFC can be used to resolve the spatiotemporal dynamics of cellular signaling. Such functionality would also enable new discoveries in applications such as studies of protein–protein interactions, ion imaging, and signaling studies.
ACKNOWLEDGMENTS
This work was partially supported by NIH grant #R43MH065724. We would like to thank Todd Kiefer, Dan Daugherty, Elizabeth Heins, and John Ritz of Prairie Technologies and Josh Bembenek and Koen Verbrugghe of LOCI for their input and contributions.
REFERENCES
- Y. Taguchi et al., "Specific biarsenical labeling of cell surface proteins allows fluorescent- and biotin-tagging of amyloid precursor protein and prion proteins," Molecular Biology of the Cell 20, 1, 233–244 (2009).
- M. Beurg et al., "Localization of inner hair cell mechanotransducer channels using high-speed calcium imaging," Nature Neuroscience advance online publication (2009).
- J.E. Linley et al., "Inhibition of M Current in Sensory Neurons by Exogenous Proteases: A Signaling Pathway Mediating Inflammatory Nociception," Journal of Neuroscience 28, 44, 11240–11249 (2008).
- J.N. Bembenek et al., "Cortical granule exocytosis in C. elegans is regulated by cell cycle components including separase," Development 134, 21, 3837–3848 (2007).
- S. Gasparini et al., "Associative pairing enhances action potential back-propagation in radial oblique branches of CA1 pyramidal neurons," Journal of Physiology 580, 3, 787–800 (2007).
Bill Vogt is chief optical engineer, Long Yan is research scientist, and Michael Szulczewski is president at Prairie Technologies, 3030 Laura Lane, Suite 140, Middleton, WI 53562-0677; www.prairie-technologies.com. Kevin Eliceiri is director of the Laboratory for Optical and Computational Instrumentation at the University of Wisconsin at Madison, 271 Animal Science Building, 1675 Observatory Dr., Madison, WI 53706-1284; e-mail: [email protected]; www.loci.wisc.edu.