ANDREW QUONG
Microscopy-based cell type assessments and genomic or proteomic analyses—recent technological advances that provide detail about cancer cell function—have enabled diagnostic and therapeutic improvements for cancer. A growing body of evidence makes clear, though, that while the genetic profile of a malignant cell is a critical factor in tumor growth, a number of other factors are equally important. These include tumor architecture and interactions among tumor cells, stroma, extracellular matrix, and immune cells.
Until recently, the inability to comprehensively interrogate these complex interactions in a spatially dependent manner has limited our ability to fully elucidate the role that tumor architecture plays in cancer diagnosis, prognosis, and therapy. The advent of Imaging Mass Cytometry (IMC), a powerful tool for spatially resolving single-cell phenotypes, has the potential to drive the next revolution in advancing cancer diagnosis and therapy.1
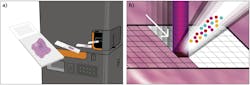
IMC, trademarked by Fluidigm (San Francisco, CA), adapts a novel laser ablation device for use with a mass cytometer (see Fig. 1).2 Tissues or fixed cell samples are stained with a panel of metal-tagged antibodies and scanned in the ablation chamber using a pulsed laser operating at 213 nm and focused to a 1-µm-diameter spot size.
Mass cytometry is then used to analyze vaporized tissue from each spot, and the metal tags present in each vaporized sample are measured and indexed against the location of each spot within the sample. Sequentially moving through the sample in a spot-by-spot basis creates a map of the sample that describes the distribution of proteins on the tissue. The map can be used for single-cell protein analysis that includes the spatial context of the cells.3
The power of IMC grows with the number of tags that can be simultaneously analyzed. Currently, there are 37 stable metal isotope labels or tags, allowing simultaneous interrogation of up to 37 targets. Data from these tags can be integrated with functional biomarker analyses and other “navigational aids” such as DNA intercalators to further orient and segment IMC images, and may enable integration of digital pathology and highly multiplexed single-cell analyses to identify novel diagnostic, prognostic, and treatment classes.3
Spatial analyses for advanced performance
Although targeted therapies have provided significant benefit in certain cancer types, the high degree of cellular heterogeneity within the tumor and surrounding microenvironment can limit these benefits if cells lack the target of the therapy, or the therapy cannot reach the cells of interest. Consequently, IMC has the potential to enable new approaches to diagnosing, classifying, and treating solid tumors based on the description of a tumor’s architecture rather than the histologic features or the molecular profile of a few of the cells present in the tumor. Two recent publications provide insight into how IMC may be used to enhance our understanding of the role genomic alterations play in cellular phenotypes and to better inform patient-specific diagnosis in breast cancer.
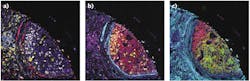
A January 2020 publication in Nature4 demonstrates how IMC can be used to identify novel breast cancer subgroups associated with distinct clinical outcomes (see Fig. 2). In this study, IMC was used to simultaneously quantify 35 biomarkers, generating 720 high-dimensional pathology images of breast tumors from 352 patients. Single-cell segmentation resulted in the identification of 855,668 individual cells in 381 images, and the expression of marker genes and spatial features was quantified for each cell. Analyses of these markers and features identified 59 tumor cell phenotypes, some of which were unique to specific patients. Further analyses identified 14 tumor cell metaclusters and 18 single-cell pathology (SCP) subgroups. Significantly, the 18 SCPs did not clearly align with classical clinical subtypes used to classify breast tumors.Comparison of SCP data with long-term survival data from 281 of the patients whose tumors were evaluated in the study found that individual subgroups had distinct clinical outcomes. As an example, one SCP (SCP 11) overlapped, but did not entirely align with the clinically assigned hormone receptor-negative, HER2-positive tumor type. Although patients in this clinical subtype typically have poor outcomes, those in the SCP 11 category had significantly better outcomes than those in the clinical subtype and other SCP groups. Similarly, the triple-negative breast cancer clinical subtype comprised five distinct SCPs, four of which had the poor outcomes associated with this subtype while patients in the fifth SCP did not succumb to the disease.
These findings highlight the prognostic limitations of current clinical subtyping strategies and underscore the potential of using multicellular spatial information generated with IMC as the foundation for a new approach to classifying and potentially treating breast cancer. Refined treatment strategies that account for cellular heterogeneity may enable improved outcomes for patients with breast cancer.
A February 2020 publication in Nature Cancer5 demonstrated that systematic analysis of single-cell phenotypic and spatial correlates of genomic alterations (mutations and copy number aberrations) provides insight into the role that the genome plays in both the composition and architecture of breast tumor ecosystems. Such insight enhances our understanding of how genomic alterations impact cellular phenotypes.
This study coupled IMC with multiplatform genomics to quantify levels of 37 proteins in 483 breast tumor samples and define the phenogenomic landscape of breast cancer. The 483 tumors evaluated in this study had previously undergone extensive genomic characterization. Clustering analysis of single-cell cell IMC expression data identified three broad phenotypic categories of tumor, stromal, and immune cells, with most cells being epithelial. The assigned cell phenotypes were validated through correlation of the proportion of cell phenotypes with overall gene expression profiles within each tumor.
Further analyses found that microRNA (miRNA), which is involved in RNA silencing and post-transcriptional regulation of gene expression, plays a more significant role in gene regulation in stromal cells compared with other cell phenotypes. The study also found unexpected differences in the phenotypic composition of tumor subtypes, as well as distinct patterns of stromal cell enrichment in different cancer subtypes. Cell-cell interactions also differed among distinct genomic tumor subtypes. Taken together, these analyses demonstrate that cellular composition and patterns of cellular interactions are highly variable across breast cancer genomic subtypes. Notably, this study also found that specific cell phenotypes were correlated with clinical outcomes, providing additional support for the potential of using IMC-based analyses to develop new approaches for stratifying patients and identifying novel therapeutic targets.
Platform for progress
Our ability to understand, diagnose, and treat cancer has always been a function of what we are able to observe. The traditional description of malignant conditions was coined about 2000 years ago based on visual observations: Cancer, the Latin word for crab, described the appearance of a tumor body and its projections, while oncology, derived from the Greek word for mass (onkos), described the tumor itself. In the subsequent millennia, much of our understanding and classification of cancer was based on what could be observed with the naked eye or the microscope, with tumors classified largely on the basis of anatomic location and gross pathology.
The advent of molecular biology not only transformed how cancer is diagnosed and treated, it also enabled powerful new tools to evaluate cancer types based on their molecular profiles, including genetic mutations and changes in the expression of key proteins in the tumor. For example, while breast and prostate cancers occur in anatomical structures with different locations and functions, molecular and genetic analyses have revealed that tumors arising in these tissues have significant biologic similarities and may benefit from similar treatment approaches. Even tumors of the same type may benefit from distinct treatment approaches based on molecular subtype classifications that allow personalized regimens.
IMC is the next step in the evolution of how we look at cancer, allowing us for the first time to see how individual cells interact with each other in the complex tumor microenvironment. As the papers described here illustrate, IMC is a powerful driver for the next oncology revolution.
REFERENCES
1. C. Giesen et al., Nat. Methods, 11, 4, 417–422 (2014).
2. R. N. Straus et al., J. Anal. At. Spectrom., 32, 1044–1051 (2017).
3. Q. Chang et al., Cytom. Part A, 91A, 160-169 (2017).
4. H. W. Jackson et al., Nature, 578, 615-620 (2020).
5. H. R. Ali et al., Nat. Cancer, 1, 163-175 (2020).
6. G. P. Risbridger, I. D. Davis, S. N. Birrell, and W. D. Tillewy, Nat. Rev. Cancer, 10, 3, 205–212 (2010).
7. M. P. Singh, S. Rai, A. Pandey, N. K. Singh, and S. Srivastava, Genes & Diseases (2019); https://doi.org/10.1016/j.gendis.2019.10.013.
Andrew Quong, Ph.D., is Chief Science Officer for Fluidigm Corporation, San Francisco, CA; e-mail: [email protected]; www.fluidigm.com.