PRM-SRS microscopy platform distinguishes specific lipid types
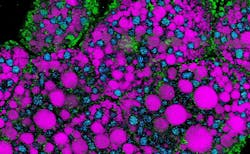
A team of bioengineers at the University of California San Diego took a major step toward better understanding lipids with an imaging platform that can distinguish lipid subtypes in even the most complex biological environments. It’s a nondestructive, label-free approach that is the first to simultaneously classify multiple lipid subtypes in cells and tissue from single label-free hyperspectral imaging sets (see video).
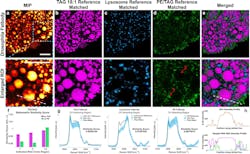
The hyperspectral imaging platform integrates a Penalized Reference Matching (PRM) algorithm, which helps to boost the accuracy in distinguishing specific lipid types, with stimulated Raman scattering (SRS) microscopy, a nonlinear method for visualizing chemical content based on molecular vibrational bonds. The combined PRM-SRS system can identify specific lipid subtypes in different organs.
Lipids—a broad group of organic compounds that includes fats and triglycerides—are difficult to see and understand accurately with commonly used methods such as fluorescence labeling. Existing technology and traditional methods require labeling the molecules of interest before imaging them with a signal that’s attached to the molecules. But rather than objective imaging, this labeling process actually alters the condition of the molecules—or can even change the physiological conditions.
“Back when I was in my postdoc training, I was always thinking about how we can use Raman spectroscopy, the Raman spectral profile, to detect the different molecules directly without any labeling,” says Lingyan Shi, an assistant professor of bioengineering at UC San Diego, whose lab developed the PRM-SRS platform. “Raman spectroscopy can be a great tool because each molecule has its own Raman spectrum profile, so you can distinguish between them without needing to label or alter anything.”
Spectral ‘soundwaves’
Shi explains that spectral profiles are akin to soundwaves. “Your voice has its own soundwave, and my voice has my own unique soundwave,” she says. “Each molecule has the same thing; each speaks in a different soundwave. That sound wave is what we consider the Raman spectrum profile—they have their own vibrational modes. The vibrational frequency ends up with a different Raman spectrum profile for each particular molecule.”
In their work with the PRM-SRS hyperspectral imaging platform, Shi’s team has been able to view and subsequently identify high-density lipoprotein particles in a human kidney as well as subcellular distributions of sphingosine, an amino alcohol that plays a role in cellular recognition, growth, and development in mammals, and cardiolipin, a phospholipid found exclusively in the inner membrane of mitochondria, in a postmortem human brain.
“Mitochondria is the cellular organelle, which is the powerhouse,” Shi says, noting it's essentially a factory that generates ATP (a small molecule that provides energy for processes in living cells, including muscle contraction and nerve impulse propagation). “The membrane has a lot of fat molecules called the carrier lutein subtype. These molecules have their own Raman spectrum profile, as well. So, no matter what kind of molecule you are investigating, they should generate a specific Raman spectrum profile that is like a soundwave for each molecule.”
Each pixel in an image has what Shi describes as a mixed spectrum that forms a final spectrum, which includes many different molecules. It’s similar to hearing numerous voices in a room—individual voices have their own soundwaves, but all together there is one collective noise. The PRM-SRS platform can detect similarities in different molecules and also differentiate between them.
“In the end, you can detect many different molecules, different fat molecules, or even other molecules like RNA and DNA, lipids, or other protein molecules,” Shi says.
Experimenting with PRM-SRS
The UCSD team’s research—published in Nature Communications—involved a look at triglycerides (dietary fats), cholesterol, and other lipid molecules, which Shi says are the most abundant molecules in certain cellular organelles.
“They’re very important to cellular machinery, which explains a lot of information—a lot of insightful information—needed for dissecting the complexity of life,” Shi says. “Our imaging platform can have far-reaching implications for human health and also scientific progress.”
The experiments revealed some surprising findings, including that different pixels in an image of the postmortem human brain tissue exhibit higher similarities than originally thought. With hyperspectral imaging, each pixel generates a Raman spectral profile. The researchers found that essentially, the Raman spectrum profile is a mixture of different molecules from the human brain tissue.
“To our surprise, as we detected each of the molecules one by one, we found that there was a high concentration of sphingolipids—a specialized group of lipids localized in the nervous system—or cholesterol in certain pixels,” Shi says. “And there were very, very high concentrations of certain fat molecules.”
Future implications
“In the past, people thought there was no way to detect fat molecules in lipids and that they could only be detected in a protein,” Shi says. “But now, we can visualize lipid molecules directly.”
The PRM-SRS platform’s ability to simultaneously visualize multiple lipid subtypes from label-free imaging sets is expanding the scope of lipid research overall. It could aid the diagnosis and prognosis of various diseases, including in the kidneys, as well as neurological disorders, and help in development of treatments.
About the Author
Justine Murphy
Multimedia Director, Digital Infrastructure
Justine Murphy is the multimedia director for Endeavor Business Media's Digital Infrastructure Group. She is a multiple award-winning writer and editor with more 20 years of experience in newspaper publishing as well as public relations, marketing, and communications. For nearly 10 years, she has covered all facets of the optics and photonics industry as an editor, writer, web news anchor, and podcast host for an internationally reaching magazine publishing company. Her work has earned accolades from the New England Press Association as well as the SIIA/Jesse H. Neal Awards. She received a B.A. from the Massachusetts College of Liberal Arts.