LIVE-CELL IMAGING: Noninvasive 3D cell imaging
Lisa Pollaro
Traditional microscopy techniques rely on invasive and complicated procedures that imply risk of damage, falsification, and error. They involve cumbersome and time-consuming preparation (taking 1–72 hours) and give rather limited results, including 2D imagery with no contrast or chemically dyed with few colors.
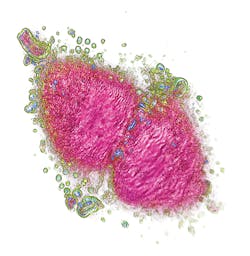
A new technology overcomes these limitations by making complete tomographic images of living cells (see Fig. 1). It does this by combining holograms obtained through a rotational scanning laser head, which illuminates the sample from 360°. Unlike computed tomography of human bodies, the technique produces the cell's tomographic reconstruction instantly, and at very low cost. The scanning laser head makes a complete rotation every second and each rotation corresponds to a 3D image, which is registered from the camera and displayed on the user's computer screen.The technique does not require expensive reagents to mark the cell parts, and the instrument itself is affordable because it does not incorporate expensive components such as fluorescent filters. As a result, users can instantly look inside the cell. The resulting image is composed of 96 z-stacks in gray scale (according to the refractive index of the cell components). The user can navigate through these layers in z. The software allows identification of the different cell parts (using a limitless number of colors), obtained instantly. By varying the transparency of the different parts, it is possible to visualize most of the internal organelles and explore the cell in 3D. A cropping plane enables "cutting away" parts of the cell to examine interior structures. It is possible to compare cells quantitatively using data collected from different experiments.
The approach enables viewing and precise measurement of the impact of stimuli and drugs on cells. For instance, the effect of a cytotoxic cancer drug can be studied on different patients' cells in order to quickly understand if the patients are sensitive or resistant to this drug. Consequently, it portends new fields of research and safer, smarter drugs.
An instrument for exploration
This approach is embodied in a new, commercially available instrument by Nanolive. The 3D Cell Explorer combines holography and rotational scanning to determine how light propagates through the cell. By this means, one can measure the refractive index distribution within the cell (3D) over time (4D). In this manner, Nanolive's microscope can be used for long-term studies, spanning up to 5 days of unperturbed observation of cultured cells in commonly available top-stage incubators. The technology enables noninvasive quantitative cell tomography and observation of nanometric details within living cells, in vitro, without sample preparation.
Holography offers a unique means to probe cells in their native environment: noninvasive and label-, manipulation-, and interference-free. Rotational scanning (with a 520 nm laser light) allows 3D reconstructions, noise robustness, and a resolution far beyond the accepted limit for light. The 2014 Nobel Prize for chemistry was awarded to S. Hell, E. Betzig, and W. Moerner, who did not believe these presumed limitations and made revolutionary discoveries in the field of fluorescence microscopy.
While their research was focused on the chemistry of single molecules and their pathways inside living cells, the new approach focuses on the physical structure of the living cell itself.
This technology allows measurement of cellular processes with real-time kinetics such as mitosis, cell movements, vesicular trafficking, and many others. It also enables multi-parameter analyses at single-cell and sub-cellular scales, such as estimations of cellular and nuclear volume. Since the cells are not manipulated in any way to introduce a label, it is possible to measure the response of a cell's physiological activity.
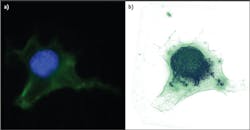
Traditionally, microscopes are handcrafted from many materials and assembled from multiple optical elements. In contrast, the 3D Cell Explorer is a completely industrialized product. Starting from a single piece of solid aluminum, the instrument's structure is milled down to include even the finest optical components. Its objective provides 60x (air) magnification using a low-power laser (λ = 520 nm, sample exposure <2 mW/mm2), with resolution of Δxy = 200 nm, Δz = 500 nm, a field of view ~80 μm, and depth of field ~30 μm. It generates a 3D image (tomograph) at ~1 frame/s.Because the average mammalian cell is 20 μm and a bacteria or yeast is 5 μm, this means we can look at subcellular structures with very high resolution and in 3D. Figure 2 shows a fixed fibroblast chemically stained to identify membrane (green) and nucleus (blue). To the right is the same cell imaged with Nanolive's 3D Cell Explorer, stained only digitally. In the first case, the preparation procedure killed the cell and took more than four hours. Using the new technology, the same result took just five minutes and would have been possible on unstained, living cells.The 3D Cell Explorer was designed for ease of use: Turn on the microscope, position the sample, and start the acquisition software. The microscope self-adjusts to the sample conditions (e.g., mounting medium or different coverslips), and within seconds, a full 3D image of the cell loads to the connected computer screen. Using the microscope's software, the user can select regions of interest and stain them digitally using any number of colors.
Software provides control
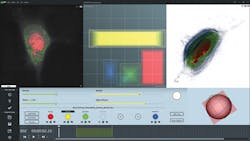
Software for Tomographic Exploration of living cells (STEVE)—a dedicated application that provides access to the microscope's features—allows, for instance, control of automatic calibration routines, 3D acquisition, and reconstruction (see Fig. 3).STEVE's intuitive graphical user interface shows all options and commands necessary for acquiring and staining images on the main screen. This window comprises three sub-windows: a viewing area that shows acquisitions as 2D slices, a 3D representation area that uses hardware-accelerated ray casting, and the Panel Viewer, which is an interactive tool for defining the digital stains applied to the acquired data.
Below the viewing areas, there is the control field, which allows access to various modes of operation. First, to quickly scan a sample, the user can choose a "white light" mode, in which the sample is illuminated using standard, incoherent wide-field illumination. The field of view is shown on the screen in real time. While the images obtained in this mode are low in contrast and limited to two dimensions, white light mode can be useful for finding a specific type of cell in a sparsely populated sample.
Once a suitable cell has been found, the user can start an acquisition. By its self-adjustment, STEVE automatically makes sure again that the microscope is properly aligned and performs calibration steps where necessary. Once running, acquisitions are performed at less than a second per 3D frame.
Digital staining, reconstruction, and processing
Unlike most optical microscopes, which measure absorption or fluorescence intensity, the 3D Cell Explorer measures the refractive index of the sample in a 3D distribution with resolution better than the diffraction limit. To mark and label certain parts of the measured cells, a process called digital staining is applied to the acquired data.
The Panel Viewer is the central control element for digitally staining a sample. It allows users to define a region in a two-dimensional space defined by the refractive index on one axis and the index's gradient norm; i.e., its spatial variation, on the other. This enables stains to be defined on regions similar to the corresponding object's refractive index and its vicinity. Stains are represented as rectangular areas in the Panel Viewer and can be modified. Changes to stains are shown in real time in the 2D view. To facilitate the digital staining process, STEVE includes a "stain painter," which users can employ to directly "paint" the part of a cell they wish to stain. STEVE will automatically define the corresponding region in the Panel Viewer.
To compute a 3D refractive index distribution from the holograms recorded in the microscope, STEVE relies on proprietary routines from the field of optical diffraction tomography. Optimized for speed and implemented to run on Graphics Processing Units (GPUs), 3D reconstruction runs in real time using affordable, consumer-grade graphics hardware.
In addition to reconstruction, STEVE uses the GPU to perform image enhancement steps based on complex deconvolution. Using this process, image aberrations (usually only avoidable by using expensive optical components) can be subtracted numerically, enhancing imaging quality and resolution. (For a free downloadable version of STEVE, please visit http://nanolive.ch/software.)
Instrumentation in practice
The first prototype of the 3D Cell Explorer was placed at the Laboratory of Lymphatic and Cancer Bioengineering (LLCB) of Professor Melody Swartz at École Polytechnique Federale de Lausanne (EPFL; Lausanne, Switzerland) in June 2014. It is used for testing cell-cell interaction between cancer, immune, and lymphatic endothelial cells. The main areas of application for the LLCB lab have been to study how immune cells interact with antigen-presenting cells, what morphological changes can be observed during this process, as well as how immune cells annihilate tumor cells and what morphological changes can be observed.
In December 2014, another prototype was placed in the laboratory of Dr. Clemens Grassberger at Harvard Medical School and Massachusetts General Hospital (Cambridge, MA).
Besides the applications described, other potential uses for 3D Cell Explorer include research on cell division, cell death (apoptosis or necrosis), and cell-cell interaction; cell morphology monitoring; cell differentiation (200+ types); intracellular trafficking; drug monitoring; in vitro fertilization; and cellular remodeling processes like neuronal remodeling before and after synapses.
LISA POLLARO, Ph.D., is communication manager at Nanolive SA, Ecublens, Switzerland; e-mail: [email protected]; www.nanolive.com.