GENETICS/DNA SEQUENCING: Laser fluorescence powers sequencing advances
DNA sequencing represents arguably the most dynamic area of bioinstrumentation by virtually any measure. Notwithstanding a 25-year history, there is increasing technology diversity (see "Profiling vs. comprehensive sequencing") rather than coalescence around a single basic approach, although laser-excited fluorescence continues to be the single most popular detection method. In fact, laser fluorescence has played a key role in the development of gene sequencing, and a fast-changing sequencing landscape is driving important product trends.
Starting with Sanger
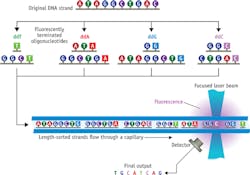
The first method to successfully sequence a length of DNA was the chain termination or Sanger method. Here, single-strand DNA is copied using a polymerase enzyme in the presence of the four different deoxynucleotide bases and a low concentration of chain terminating nucleotides; the latter are chemically modified so that the enzyme can incorporate one of these nucleotides, but their structure prevents any further synthesis. The result is a mix of every length from the original so-called primer sequence of a few bases to the entire original strand length. Through radioactive labeling of the terminating nucleotides and spreading the copies across a plate of silica gel, the sequence could be read as the characteristic four-track (A, C, G, T) "bar code" on an exposed photographic plate.
First-generation instruments automated this method by switching to fluorescently labeled, chain-terminating nucleotides (a different fluorescence signature for each of the four bases) and separating the various copy lengths via capillary electrophoresis (CE) followed by (typically 488 nm) laser-excited fluorescence to distinguish among A, C, G, and T (see Fig. 1).
Each run could sequence just a few hundred bases at a time. So, the entire human genome was read by multiple labs, each operating multiple sequencers—with a total cost of around $3 billion over 10 years.
Next-gen: Massive parallelism
Incredibly, the cost to sequence an entire human genome has now dropped by nearly seven orders of magnitude to under $1,000 per complete human genome. And developers of third-generation instruments are even touting a further possible 10-fold cost reduction.
The massive parallelism used in next-generation sequencing (NGS) instruments was a vital step that enabled this progress in speed and cost. The two most popular NGS methods proved to be those developed by Illumina (San Diego, CA) and Life Technologies (Carlsbad, CA). Both rely on first cutting the target DNA into manageable strands of typically a few hundred bases. These are then arranged in some type of non-overlapping array and amplified in situ—by polymerase chain reaction (PCR) or cloning—to form small clusters that each contain a different strand, but many identical copies of that strand. The sequencing is followed fluorescently using a multi-megapixel sensor so that laser fluorescence from tens and even hundreds of thousands of clusters can be analyzed simultaneously. Life Technologies uses sequencing by ligation (cutting), whereas Illumina relies on sequencing by synthesis (SBS). The latter has become the market leader, in part because the incredible throughput of their flagship instrument at up to 600 billion bases per day makes it well-suited for applications like whole-genome sequencing.
In both cases, the sequencing is carried out on a glass plate or flow cell excited with laser light. Each of the four nucleotides (A, C, G, and T) has a unique fluorescence emission profile. So each unique cluster (i.e., image location) flashes a wavelength pattern (color) corresponding to the next base to be cut (Life Technologies) or incorporated (Illumina). The combination of dichroic filters and low-noise digital camera detection enables the sequence of these millions of clusters to be simultaneously read, base by base, with each chemical cycle. The computer then compares all the sequences from these randomly arranged clusters, and often compares these to known/expected genome sequences to assemble the entire sequence of the original uncut DNA.
Third generation: Technological diversity
Looking to the future, there are numerous "third-generation" sequencing methods in various stages of commercial development, including new electrochemical and semiconductor-based detection systems. For several reasons, however, laser-excited fluorescence is the dominant method: It is well proven and also the most versatile way of sampling wet chemistry and biochemistry in real time, with unmatched spatial resolution and high (even single-molecule) sensitivity.
The goals of most third-generation methods are lower costs and even faster overall throughput. We emphasize the term overall because several factors beyond speed—bases per run, strands per run, and strand length, for instance—determine throughput. Whereas NGS methods feature myriad short reads, third-generation approaches typically emphasize just a few very long reads. This simplifies the software challenge of overlapping the oligonucleotide strands to assemble the complete target sequence, reducing the necessary computer time and reducing the amount of oversampling (sampling depth).
Oversampling is necessary because NGS and some third-generation methods often have high error rates. Unlike the near 100-percent accuracy of modified Sanger, some techniques deliver error rates (per individual read event) of 15 percent or more. However, providing the errors are truly random; then, the oversampling that is built into these methods can still deliver essentially 100-percent accuracy. For example, even if each base for each read delivers just 80-percent accuracy, then 15–30 times oversampling yields infinitesimally small final error rates.
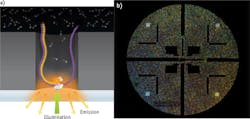
One particularly interesting method with novel optics has been developed by Pacific Biosciences (Menlo Park, CA). The PacBio RS II system uses zero-mode waveguides (ZMWs)—optical waveguides whose diameter is much smaller than the wavelength of light. On a so-called Single Molecule, Real-Time (SMRT) chip, ZMWs are created by patterning circular holes (approximate diameter 70 nm) in an aluminum film (100 nm thickness) deposited on a clear silica substrate. A single polymerase enzyme is located at the bottom of each waveguide. Laser illumination occurs through the silica and can penetrate only about 30 nm into each waveguide, so millisecond pulses of fluorescence can only be efficiently collected from the nucleotide as they are being added to the DNA. Importantly, the company claims single, continuous read lengths as long as 40,000 bases. The utility of a sequencing instrument is determined by the degree of parallelism that can be reached in scaling up this simple experiment, and this comes down largely to available laser technology. In the PacBio RS II, 150,000 ZMWs are monitored simultaneously, in real time, by dividing the lasers into that many individual diffraction-limited spots targeted to the individual ZMWs (see Fig. 2). According to Paul Lundquist, Senior Manager of Optical Engineering and Instrument Design at Pacific Biosciences, Coherent's Genesis technology was well suited for this application. "It is not difficult to generate a diffraction-limited spot from a laser beam, but the optical system required for this level of beam splitting and precise positioning required very good beam quality and wavelength stability. These capabilities and the scalable power platform made the Genesis a good fit."At least in the short term, these third-generation technologies will compete with NGS, so it is worth noting a couple of interesting photonic advances to the dominant NGS techniques just described. One development is to attach the oligonucleotide strands to small (3 μm) glass beads, rather than randomly on a surface. After bridge amplification, these are then self-assembled on the lower surface of a flow cell with a dense array of small pits or wells. This ensures the highest possible density while still avoiding overlap of the clusters or beads. Moreover, using microelectromechanical systems (MEMS) techniques, the outer lower surface of the flow cell incorporates a pattern of near-parabolic reflectors that concentrate laser light on each bead and also efficiently collect epifluorescence. Laser light is then efficiently focused into these optical collectors using a microlens array. Another development is in the fluoro-chemistry that simplifies the optical system and reduces the total number of images required. Specifically, some systems use a two-dye sequencing chemistry and two-channel detection, where 'C' is red-only, 'T' is green-only, 'A' is a mix of red and green, and 'G' is "dark" (no red or green).
Laser performance, integration
All this impressive innovation in chemistry, nanophotonics, and microfluidics drives four key trends in lasers: diversity, reliability, integration, and partnering.
Laser diversity. The dynamic nature of sequencing and the nascent state of the end market—clinical applications currently represent only a few percent of the market—make the future hard to predict. Only a handful of wavelengths (488 nm, 514 nm, 532 nm, and red-say, 640 or 660 nm) are required, but diverse choices are needed in every other laser parameter. This means offering the widest possible choice of standard lasers to support R&D, which in turn often requires more than one core technology (e.g., diode, optically pumped semiconductor laser [OPSL]). It also means being able to create cost-effective custom lasers to support optimized, higher-volume instruments.
For example, Coherent supplies the market with laser powers ranging from a few milliwatts to 10 W. Why? For applications where the DNA strands are highly amplified and spatially concentrated, the fluorescence signal is more intense and less laser power is required. But if these sampling sites are then spread over large flow cells or slides needing wide field illumination, that will increase the power requirement. For applications where single strands are being sequenced and signal is therefore inherently low, power is critical, particularly as these applications usually feature massive parallelism. Another factor in the power equation is damage; the laser must not cause functional damage to any of the enzymes used in the sequencing, or to the DNA itself. And because shorter wavelengths are generally more damaging, there is less of a demand for power above a few hundred milliwatts at shorter wavelengths like 488 nm.
Laser reliability. While most commercial laser applications require reliability, the issue here is not just the cost of unscheduled downtime for instrument OEMs and their end users. Some of the sequencing technologies, particularly those targeted at full genome applications, are designed to run continuously for up to several days. If the laser performance fails or even momentarily drifts during that time, the entire set of data can be compromised and rendered useless. As a result, the market is very conservative and not interested in unproven laser technologies.
Laser integration. For most sequencing techniques, the proprietary and patented "magic" and consumables-dominated profit stem from the patented chemistry, microfluidics, and so forth: there is less innovation in how the laser light is used. Plus, most of these companies have no investment or expertise in how the laser light is generated and delivered/modified. So, in most cases, they are looking for a complete photonic engine rather than a standard laser. During R&D and product breadboarding, at a very minimum instrument developers need smart lasers with plug-and-play simplicity. But they also often need off-the-shelf modular solutions to enable fiber delivery (both single- and multimode), plug-and-play combining of multiple wavelengths, and beam shaping using refractive and diffractive optics (see Fig. 3). And in some cases, they require an engine in which the coherence of the laser is eliminated in order to avoid speckle and ensure uniform stable illumination.Partnering. As a result of these market characteristics, instrument developers and manufacturers are typically looking for laser suppliers to act as subcontractors rather than as component vendors. This partnering includes the capability to create systems to deliver certain output specifications, as well as—less commonly—systems built to print. In addition to this expertise, instrument companies often look for a laser sub-contractor that can support unpredictable growth patterns as these different third-generation sequencing technologies are successfully commercialized and begin to compete in a fast growing market.
Adding value
Despite huge technological advances, sequencing still represents a dynamic nascent market characterized by incredible technological diversity. Moreover, the laser is viewed as a workhorse component, not an innovative solution. This means that laser manufacturers have to support this industry with a diverse family of off-the-shelf lasers and beam delivery/beam combining products, as well as the ability to provide custom systems with volume scaling.
Profiling vs. comprehensive sequencing
A varied market of applications is one of the reasons for growing technological diversity in sequencing methods. Clinical applications constitute a small fraction of the DNA sequencing market, though significant growth (nearly 2X) is predicted through 2019 (proprietary research). Moreover, only a few research and clinical applications require reading gigabases at single-base resolution, as in the complete sequencing of the first human genome. Some applications—for instance, looking for spot mutations associated with certain cancers or developing genetically modified crops—need single-base resolution of short sections.
In profiling applications, the goal is to look for the presence and/or location of certain sequences or genes rather than reading every single base. An interesting method by BioNano Genomics (San Diego, CA) combines novel microfluidics with laser-excited fluorescence. The method inserts fluorescently labeled strings of single-strand DNA into the double-helix DNA being profiled. Each string is inserted only where there is an exact complementary match. The DNA is then forced through a long, narrow microfluidic channel, which stretches out the DNA tertiary structure to create a long, string-like shape. This flow is then intersected with a laser source and the inserted sections of DNA can be read like a color bar code.
Volker Pfeufer | Senior Product Line Manager, Coherent GmbH
Volker Pfeufer is Senior Product Line Manager for Diode-pumped Solid State Lasers at Coherent GmbH (Luebeck, Germany).
Matthias Schulze | Director of Marketing, OEM Components and Instrumentation at Coherent
Matthias Schulze is Director of Marketing, OEM Components and Instrumentation at Coherent (Santa Clara, CA).