New brightness-equalized quantum dots allow for improved bioimaging
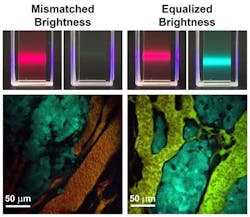
A new class of light-emitting quantum dots (QDs) with tunable and equalized fluorescence brightness across a broad range of colors results in more accurate measurements of molecules in diseased tissue and improved quantitative imaging capabilities, thanks to work by researchers at the University of Illinois at Urbana-Champaign.
Related: Silicon quantum dots promising for clinical deep-tissue imaging
Andrew M. Smith, an assistant professor of bioengineering who led the work, explains that light emission had an unknown correspondence with molecule number previously. But now, it can be precisely tuned and calibrated to accurately count specific molecules, which will be particularly useful for understanding complex processes in neurons and cancer cells to help unravel disease mechanisms, and for characterizing cells from diseased tissue of patients, he says.
Sung Jun Lim, a postdoctoral fellow and first author of the paper describing the work, says that it has always been challenging to extract quantitative information because the amount of light emitted from a single dye is unstable and often unpredictable. Also, brightness varies drastically between different colors, which complicates the use of multiple dye colors at the same time. These attributes obscure correlations between measured light intensity and concentrations of molecules, he adds.
The new materials will be especially important for imaging in complex tissues and living organisms where there is a major need for quantitative imaging tools, and can provide a consistent and tunable number of photons per tagged biomolecule, the researchers say. They are also expected to be used for precise color matching in light-emitting devices and displays, and for photon-on-demand encryption applications. The same principles should be applicable across a wide range of semiconducting materials.
Full details of the work appear in the journal Nature Communications; for more information, please visit http://dx.doi.org/10.1038/ncomms9210.
Follow us on Twitter, 'like' us on Facebook, connect with us on Google+, and join our group on LinkedIn