Light microscope can image whole living organisms at high resolution
Researchers at the Howard Hughes Medical Institute (HHMI)'s Janelia Research Campus have developed a new light microscope that gives a clearer, more comprehensive view of biological processes as they unfold in living animals. The microscope produces images of entire organisms, such as a zebrafish or fruit fly embryo, with enough resolution in all three dimensions that each cell appears as a distinct structure. What's more, it does so at speeds fast enough to watch cells move as a developing embryo takes shape and to monitor brain activity as it flashes through neuronal circuits.
Philipp Keller and his team at Janelia aim to understand how a functioning nervous system emerges in an embryo. Over the last five years, they have devised several imaging technologies that make it possible to image large biological samples at high speed. His lab’s newest microscope, called the IsoView light-sheet microscope, overcomes a final challenge—improving spatial resolution—without sacrificing the performance features of his team’s previous microscopes. The IsoView microscope is described in a Nature Methods paper, and includes complete building plans for the microscope and the associated image processing software developed by the team.
In 2012, Keller's team developed the SiMView microscope, which provides fast 3D imaging of large specimens. His lab has used the technology to follow cells for days as they move and divide throughout entire living embryos, and to watch individual neurons fire throughout an entire central nervous system. Still, structures inside cells seemed blurred together, and cells deep inside a sample could not be seen well at all.
In developing the IsoView microscope, Keller's team sought to overcome a limitation inherent in light microscopy: no matter how good spatial resolution is along the two lateral dimensions of an image, resolution is much poorer along the third axial dimension. Keller explains that this is because of the way a microscope's objective lens collects the light emitted from a sample. That light, which is used to produce the image, projects every which way, but a microscope's objective can only detect photons that fall within a small cone; information from outside that cone of light goes unused. The result, Keller explains, is that spatial resolution along the detection axis is always much worse than it is along directions perpendicular to the detection axis.
Scientists have found ways around this problem, and several new imaging technologies produce astonishingly detailed 3D images. But none of those technologies combines high spatial resolution with the other features that Keller needed for his experiments. Rather than collecting a single image of a sample with a single objective, the new microscope simultaneously collects light and creates images from multiple angles. Each image still suffers from poor resolution along one axis, but the most useful data from each image can be combined to generate a final image with good resolution in all dimensions.
Keller and a postdoctoral researcher in his lab, Raghav Chhetri, designed a microscope with four objective lenses positioned around the sample at right angles to one another. Each objective sends light into the sample to illuminate it, and also collects fluorescent light emitted by the sample.
From each side, an objective produces a thin beam of light that sweeps the sample from top to bottom so quickly that the detection camera across from it sees a continuous sheet of light. The beams from each of the four objectives are staggered so that they do not interfere or intersect with one another, and a rolling shutter in each camera keeps pace with the beam, so that the camera's detector remains focused on the narrow slit along which it can pick up high-resolution information. These features allow the microscope to collect four different images of the sample from different angles simultaneously, without any crosstalk between these multiple views.
Computer scientist Fernando Amat developed image-processing software to transform those images into a final high-resolution image. Keller notes that there was no software available that could do the job—it required developing an entirely new software package that could speed image-processing strategies by more than 60-fold. A typical experiment with this microscope might run for one hour. During that time, the four sCMOS cameras in the microscope can stream data from the experiment at rates up to 3.2 Gbytes/s. This results in about 10 Tbytes of multi-view image data from one hour of imaging. With the new algorithms, an IsoView dataset can be processed in about two days with a single workstation.
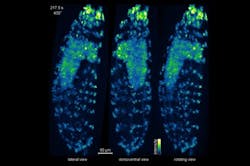
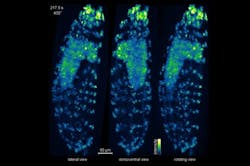
Keller and his colleagues have used the IsoView to visualize cell-by-cell activity throughout the nervous system of an entire living fruit fly larvae, an organism that has more than 10,000 neurons and is about 50 times larger than the roundworm C. elegans, the only animal whose complete nervous system has previously been imaged at the single-cell level. Because the IsoView can produce images as the larvae moves freely in a loose gel, Keller says, “this opens up the possibility of functional imaging in an entire, behaving animal. It is even possible to perform high-speed functional imaging over developmental time scales, as we demonstrated in imaging a fruit fly embryo developing into a larva.”
The scientists also performed high-resolution functional imaging of activity in the entire brain of a larval zebrafish, demonstrating that neurons in the deepest, least accessible regions of the brain could be seen clearly, separate from their neighbors. Finally, they used the IsoView to track cells in a developing fruit fly embryo. A multicolor imaging mode enhanced the information presented in the resulting videos by detecting different fluorescent labels on cells' membranes and nuclei.
The team plans to continue improving the IsoView technology, but Keller is also eager to begin using it for biological experiments.
Full details of the work appear in the journal Nature Methods; for more information, please visit http://dx.doi.org/10.1038/nmeth.3632.
Follow us on Twitter, 'like' us on Facebook, connect with us on Google+, and join our group on LinkedIn