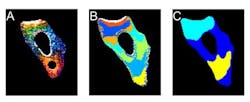
Fluorescence microscopy shows how unfolded proteins move in the cell
Using a fluorescence microscopy method, University of Illinois (Champaign, IL) researchers were able to track how an unfolded protein diffuses in a cell. Studying the relationship between protein folding and transport could provide great insight into protein-misfolding diseases such as Alzheimer's and Huntington's.
Related: UMass researchers use FRET to study protein misfolding diseases
"We're looking at the earliest stages of disease, the initial phases of transport of bad proteins," explains chemistry professor Martin Gruebele. In the past, he says, much research on Alzheimer's and similar disease focused on fibrils—large bundles of misfolded proteins that form in the brain.
"But now, we think the fibrils are just an end product that's left over when the cell dies, and the actual killing mechanism has to do with migration of the protein to specific places in the cell like the outer membrane," Gruebele says. "Understanding how these mechanisms work at a fundamental level is going to give people more handles on where to look to cure things."
Researchers have hypothesized that an unfolded protein moves more slowly through the cell, because it would be a big, stringy mess rather than a tightly wrapped package. So, the Illinois researchers devised a way to measure how diffusion slows down when a protein unfolds using a fluorescence microscope, and then used 3D diffusion models to connect the protein's unfolding to its motion.
The researchers found that the unfolded protein did indeed slow down, although its speed was not steady. It sometimes zoomed swiftly to a new location, and sometimes sat idling in one area. They were able to map out areas of the cell with different rates of diffusion, the cellular version of a speed limit.
The unfolded protein's slowdown is not only due to size, however. The researchers did additional experiments to prove that the unfolded protein stuck to other molecules in the cell. A class of molecules in the cell called chaperones have the job of binding to parts of proteins that come unfolded, and the researchers found that the unfolded protein interacted more with chaperones than did the properly folded protein. However, when high numbers of proteins unfold, the cell’s systems can get overloaded and the chaperones can’t handle them all.
"Looking at something like this can start to give people a handle on why something that seems relatively harmless in vitro sometimes can have such a large effect in the cell," says Hannah Gelman, a graduate student who was involved in the work. "A change that makes a slightly less effective protein in the test tube can turn into a completely fatal mutation in the cell. First, the protein's role in the cell can no longer be fulfilled. Second, as more and more unfold, they can disrupt the function of the whole cell."
The researchers think that the unfolded protein is likely to stick to nonchaperone molecules as well, causing other problems in the cell and disrupting the flow within a cell. They plan to use the fluorescence microscope to study other proteins and how unfolding affects their diffusion, to see if the properties they observed are universal or if each protein has its own response.
They also hope to use their method to watch how unfolded or misfolded proteins move to the cell's membrane, where they aggregate and create the problems seen in Alzheimer's and other diseases.
Full details of the work appear in the journal PLoS One; for more information, please visit http://dx.doi.org/10.1371/journal.pone.0113040.
-----
Follow us on Twitter, 'like' us on Facebook, connect with us on Google+, and join our group on LinkedIn
Subscribe now to BioOptics World magazine; it's free!