DNA signal-amplification technique visualizes several proteins within one sample
Seeking to better understand how tissues and organs develop, fail to function, and regenerate over time, a team of researchers at Harvard University's Wyss Institute for Biological Engineering and Harvard Medical School (HMS; both in Cambridge, MA), led by Peng Yin, Ph.D., has developed a DNA nanotechnology-based approach they call Immunostaining with Signal Amplification By Exchange Reaction (Immuno-SABER). The method combines the protein targeting specificity of commonly available antibodies with a DNA signal-amplification strategy that enables the highly multiplexed visualization of many proteins in the same sample with pre-programmable and tunable fluorescence signals at each target site. The team has validated their method in a broad range of cell and tissue preparations.
"We demonstrated that Immuno-SABER provides the capability to independently tune the signal intensity for individual protein targets 5- to 180-fold, with multiplexing capability to allow the simultaneous detection of many proteins. Together with its speed, relative ease of use, and low costs, this technique has the potential to fast-forward ongoing large-scale protein-mapping studies and biomarker discovery efforts across many tissues and diseases," says Yin, who is a Wyss Institute Core Faculty member.
Related: Fluorescence analysis method boosts DNA and RNA visualization
Antibodies are the most common detection reagents for proteins both in research and clinical settings. They are typically tagged with fluorescent stains to make them detectable by microscopy. However, conventional antibody staining methods typically allow only a maximum of five different stains to be used simultaneously, and target proteins can differ significantly in their abundances, making it difficult to distinguish rare protein targets with high sensitivity from the background fluorescence that many tissues display.
Immuno-SABER, however, utilizes the "Primer Exchange Reaction" (PER) method previously reported by Yin's group to synthesize long concatemers of short DNA primer sequences with the help of a catalytic DNA hairpin structure. The PER-generated concatemers are attached via short handle sequences to DNA-barcodes on antibodies that bind to target proteins in fixed cell and tissue samples with high specificity. At the target site, SABER concatemers provide a scaffold with multiple binding sites for complementary fluorescent oligonucleotides ("imagers"), and thus a means to amplify the signal emanating from each protein target.
"By barcoding antibodies with unique short DNA sequences and applying Immuno-SABER, we can simultaneously visualize multiple protein targets on the same sample and with high specificity. This essentially opens up a way to analyze the protein variety present in tissues in a robust and multiplexed fashion," said co-first and co-corresponding author Sinem Saka, Ph.D., who works as a Postdoctoral Fellow on Yin's team.
The team significantly boosted the multiplexing potential of their Immuno-SABER approach by coupling it with their previously developed "DNA-Exchange" technique. In DNA-Exchange, imagers that mark one set of target proteins are washed off and replaced by another set of imagers marking a different group of target proteins and this can be repeated multiple times.
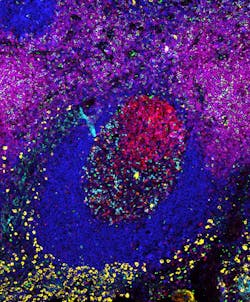
Previously developed methods for highly multiplexed protein detection that work by repeating some of their key steps at different protein targets tend to suffer from suboptimal sensitivities, or take considerable time (low throughput) and finesse to execute. "Exchange-SABER provides high sensitivity with one single step of staining and amplification, and high multiplexing and throughput with multiple fast imager exchange steps," says Yu Wang, a graduate student on Yin's team and co-first author of the paper that describes the work. "As proof-of-concept, we visualized 10 different proteins in cryosections of the mouse retina."Another key facet of Immuno-SABER facilitating the parallel detection of many proteins at a time is its ability to tune signal strength. The research team achieved this by assembling more complex branched structures from PER-generated concatemers that contain higher numbers of binding sites for fluorescent imagers. "Programming the complexity of PER-based concatemer structures allows us to tune the signal strength to the abundance of particular proteins. We can at the same time visualize rare proteins with branched SABER products that enable higher signal amplification, and abundant proteins with linear SABER products," says Saka. In their study, the team combined linear and branched SABER concatemers to, for example, simultaneously visualize six protein targets with different abundances and cellular locations in human tonsil samples.
To achieve similarly high resolution of proteins in more complex tissue environments, the team combined Immuno-SABER with a method known as expansion microscopy, which was previously developed by co-author Edward Boyden, Ph.D., the Y. Eva Tan Professor in Neurotechnology at MIT. The expansion method swells fixed tissues artificially to larger volumes, which increases the separation distance between individual molecules and thus improves their effective resolution without the need for specialized instruments.
"Combining expansion microscopy with Exchange-SABER simultaneously gives us the high-multiplexing, -throughput, and -resolution capabilities needed to move efforts such as building molecular atlases for the human body more effectively forward,” says Wang.
Full details of the work appear in the journal Nature Biotechnology.