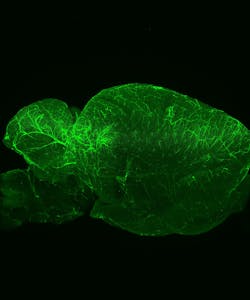
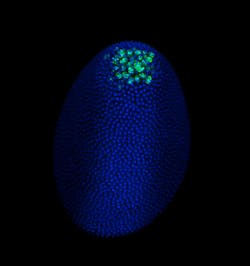
FLUORESCENCE MICROSCOPY/CELL BIOLOGY: The planar truth about light-sheet microscopy
New technologies can set off leaps in biological imaging. One of the most recent comes from light-sheet fluorescence microscopy (LSFM), a technique that optically sections a specimen with speed and collects data for three-dimensional reconstructions. Biologists can optically section a sample without LSFM by focusing deeper into it, but that approach requires extensive manual repetition and illuminating the entire sample throughout the process, which damages some tissues. LSFM, on the other hand, only illuminates the sample slice by slice. Although LSFM usually provides only intermediate resolution, that can be enhanced with other forms of microscopy. For example, a team of Italian scientists combined one form of LSFM called selective plane illumination microscopy (SPIM) with a super-resolution technique, and reported localizing single molecules in live samples.1 Other work shows the appeal of SPIM in developmental biology.2 Even in conventional setups, LSFM meets the needs of many applications in cellular biology. In addition, it offers the field of view and sample conditions to image large samples, including living embryos. Experts from three organizations—Carl Zeiss Microscopy (Oberkochen, Germany), Harvard University (Cambridge, MA), and LaVision BioTec (Bielefeld, Germany)—explained the technology behind LSFM and revealed the growing list of applications.
Although some of today's most advanced imaging technologies require on-site support, Douglas Richardson, director of imaging at the Harvard Center for Biological Imaging, points out that making images with LSFM can be easier than expected. "Light-sheet is great, as it's easy to sit down and bang out good images quickly," he says.
But success with LSFM requires that the scientist understand the unique aspects of this type of microscopy. Other microscopes generally use epi-illumination, in which the light and detection come from the same side of the sample. In addition, most microscopes use the same lens for illumination and detection. With LSFM, the illumination comes at 90° to the detection, and the platform uses different lenses for the processes. So, as Richardson says, "At first, it's a bit of a geometry quiz to wrap your head around." In addition, LSFM, as the name suggests, uses a sheet of laser light for illumination. Most important, this sheet illuminates only the part of the sample that is being observed. "This dramatically reduces photo damage," says Scott Olenych, product marketing manager for imaging products at Carl Zeiss.
In addition to the differences in the optics, this technology captures images with a CMOS camera, making it possible to "get through a z-stack very fast," Olenych says. "It collects the entire plane at once, about 2000 × 2000 pixels."
Spreading samples
At Harvard, Richardson expected to apply LSFM to Drosophila and zebrafish embryos, primarily, but the applications turned out to go far beyond those organisms. "That's maybe one quarter of what we are doing now," he says. It's pretty simple to use, he says, and "lots of people like it for larger fixed samples—everything from CLARITY-cleared brains to mouse embryos." LSFM also works for a range of live samples, from model organisms like embryos of the commonly studied roundworm C. elegans embryos, to less expected samples, like salamander embryos. Richardson's colleagues image those samples and more. If the sample is 5 mm or less in size, it might work in LSFM using the Zeiss platform.
To see below the surface, though, the sample must be optically clear. In a CLARITY-cleared mouse brain, for example, Richardson says that LSFM can image through one that is up to 5 mm thick. Something that is naturally transparent doesn't need any clearing.
Zeiss's Lightsheet Z.1, the first commercially available LSFM system, gives biologists another option for seeing more. A sample is suspended in front of the detection objective, and it can be rotated for imaging from all sides. "Maybe you can look at a sample from one side and see to a certain depth, then image from the other side for more," Olenych says.This technology improves imaging for many biological samples, but it does not provide the best imaging for every situation. "The only samples we don't recommend are things like thin mounts of cells," Olenych says. With such specimens, "you get better resolution and cell stability from another type of system, like a spinning disk."
Even more biologists will turn to LSFM as additional commercial platforms become available. In addition to the Lightsheet Z.1, scientists can consider the Ultramicroscope from LaVision BioTec. Uwe Schröer, product manager at LaVision BioTec, describes this LSFM platform as "better than micro-computer tomography with a larger field of view than conventional microscopes." He adds, "It lets you see connections and see how systems work—how populations of cells are connected." Beyond using this platform in developmental biology and for larger samples, Schröer says that researchers have imaged the vascularization of tumors.
Dealing with the data
The right software makes all of the difference in using LSFM. For one thing, software continuity doesn't hurt. As Richardson says, "We're lucky here in that every system we have is Zeiss, and they all run ZEN [(Zeiss efficient navigation) imaging software]. So everyone here already knows it, which makes it easier to jump into LSFM."
Beyond the continuity, Richardson appreciates variety on how software can be used. He says, "The one thing that I notice about Zeiss software is that there is a checkbox or knob or button to do everything, but ZEN lets you choose how much of that you let users see." Then, the system can be adjusted for beginners and experts, giving the latter group more control.
"With LSFM, the biggest issue is dealing with the sheer amount of data that you get," Richardson says. Some researchers already generate terabytes of data—that's 1,000 Gbytes. "Potentially, you may need a computer that can hold tens of terabytes of data in memory in order to do certain types of downstream processing," he says. In some cases, special tools are already being developed for LSFM. For example, scientists are creating software able to use less memory when opening big images.
The size of the image, though, depends on how a biologist uses LSFM. For example, Schröer says, "If the process does not include time-lapse images, then the files are usually only 5–10 Gbytes."
Despite the data complexity, many scientists can quickly use LSFM. According to Olenych, "We have people collecting data very rapidly." That's all with very little training, at least for someone experienced in other forms of biological imaging. He adds, though, that "processing the images is more complicated and that takes more time." LSFM also requires the right computer. Olenych says, "You need a decent amount of RAM on a workstation."
Even with the large amounts of data creating challenges, LSFM makes many biologists smile. As Richardson says, "Nearly everyone who's tried it is super excited about it."
References
1. F. C. Zanacchi, et al., Nat. Meth., 8, 1047–1049 (2011).
2. J. Huisken and D. Y. R. Stainier, Development, 136, 1963–1975 (2009).
Mike May | Contributing Editor, BioOptics World
Mike May writes about instrumentation design and application for BioOptics World. He earned his Ph.D. in neurobiology and behavior from Cornell University and is a member of Sigma Xi: The Scientific Research Society. He has written two books and scores of articles in the field of biomedicine.