STIMULATED RAMAN SCATTERING MICROSCOPY: Vibrational microscopy method generates high-res images of nascent proteins in living cells
Researchers at Columbia University (New York, NY) report a method that enables something never before achieved: selective, high-resolution visualization of newly synthesized proteins in living biological systems.1 The breakthrough has broad implications because the ability to directly visualize and quantify budding proteins at a global (i.e., proteome) level is extremely valuable for understanding the metabolic activities of living cells. Such spatio-temporal characteristics of protein synthesis are critical determinants in such intricate biological processes as cell growth, differentiation, disease, and stimulus response. For instance, long-lasting forms of synaptic plasticity, such as those underlying long-term memory, require new protein synthesis in a space- and time-dependent manner. In terms of neurological diseases, excessive synaptic protein synthesis has recently been proposed as one mechanism contributing to autism (a devastating neurodevelopmental disorder) in humans.
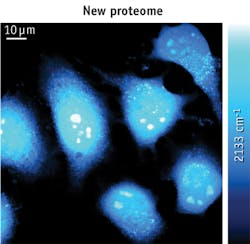
The Columbia team's technique involves stimulated Raman scattering (SRS) microscopy, along with metabolic incorporation of deuterium-labeled amino acids. By detecting the unique vibrational signature of carbon-deuterium (C-D) bonds carried by the amino acid building blocks, the technique allows visualization of newly synthesized proteins and clear identification of subcellular compartments with fast protein turnover (e.g., in HeLa and HEK293T cells), and differentiation of newly grown neurites in neuron-like N2A cells. As a first demonstration, the researchers created high (spatial-temporal) resolution images of nascent proteins in live mammalian cells.
According to Lu Wei, the lead author of the study, "our work represents the first time that nonlinear vibrational microscopy has been used to visualize the metabolic incorporation of isotope labeled precursors of macromolecules." She adds that incorporation of deuterium-labeled amino acids is minimally perturbative to live cells, and SRS imaging of exogenous C-D bonds in the cell-silent Raman region is highly sensitive, specific, and compatible with living systems under physiological conditions—without requiring fixation or staining.
Assistant Professor Wei Min, who led the work, notes that the biocompatiblility of deuterium labeling, along with SRS imaging, enables the possibility of monitoring protein synthesis in more complex systems such as live tissues and animals. He is currently working towards this exciting goal, and foresees that the technique "will be a valuable imaging tool for biomedical researchers to probe the complex spatial and temporal dynamics of proteomes in vivo."
1. L. Wei, Y. Yu, Y. Shen, M. C. Wang, and W. Min, Proc. Nat. Acad. Sci., 110, 28, 11226–11231 (2013).