Tweaked enzyme that produces light could improve medical diagnostics
With the goal of a low-cost, simple, and highly accurate detection system that could change medical diagnostics, École Polytechnique Fédérale de Lausanne (EPFL; Switzerland) scientists have chemically tweaked the enzyme responsible for the light of fireflies to make it seek target biological molecules and give out a light signal.
Related: Breath test that contains silicon microsensors can detect cancer
The lab of Kai Johnsson at EPFL, led by Alberto Schena and Rudolf Griss, were able to add a small chemical tag on the enzyme luciferase, which produces the light of fireflies. The tag detects a target protein, and the luciferase gives out a light signal that can be seen with a naked eye.
In 2014, the research team developed a quick and easy drug-monitoring molecule that led to a startup company, Lucentix. Thinking outside the box, they bypassed the pains of protein engineering altogether: instead of mutating the luciferase to make it sensitive for a target protein—which would require enormous labor—they simply attached it to a small chemical tag.
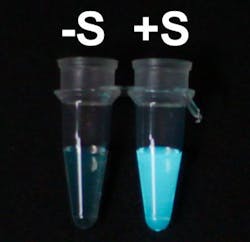
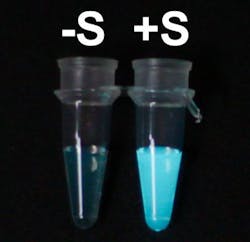
The tag acts as a switch by blocking luciferase, thereby preventing it from producing light. When the tag detects its target protein, it attaches to that instead, removing the block from lucifarase. As a result, luciferase is free to turn on the lights, which is the signal that the target has been found.
"You can think of the tagged luciferase as a cyborg molecule," says Johnsson. "Half bio, half synthetic. How could you make luciferase sensitive to the presence of another protein just through mutations? It's a lot of work. With this chemical trick, all we have to worry about is designing an appropriate tag that can recognize the target protein."
The activation of luciferase when it detects its target protein is dramatic enough to see with a naked eye. This means that the system does not demand expensive and complicated readout devices.
Full details of the work appear in the journal Nature Communications; for more information, please visit http://dx.doi.org/10.1038/ncomms8830.
Follow us on Twitter, 'like' us on Facebook, connect with us on Google+, and join our group on LinkedIn