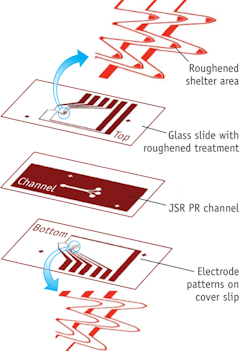
SERS-based microfluidic chip distinguishes bacteria
Engineers at Taiwan's National Cheng Kung University have created an on-chip method for sorting and identifying bacteria.1 The surface-enhanced Raman spectroscopy (SERS)-based technique uses roughened glass slides patterned with gold electrodes, which the researchers used to create microchannels for sorting, trapping and identifying the bacteria. Proteins or other chemical components on the surface of bacteria become attached to the craggy gold zone; when excited, these components cause representative peaks at different wavelengths, creating spectral "fingerprints." Although some species of bacteria could show very similar signatures because the components on their surfaces are almost the same, bacteria from different genera are distinguishable using the technique.
By optimizing conditions such as the flow rate, applied voltage, and frequency, it is conceivable that the same approach could be used to sort different species of fungi, on the basis of their different electrical or physical properties. Thus, a portable device could enable preliminary screening for pathogenic targets in bacteria-infected blood, and in raw milk, for example.
1. I.F. Cheng et al., Biomicrofluidics 4: 034104 (2010)
More BioOptics World Current Issue Articles
More BioOptics World Archives Issue Articles