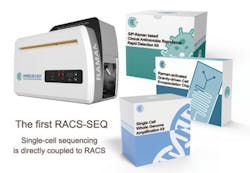
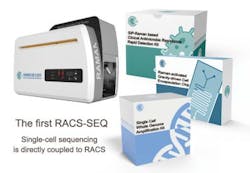
China develops Raman-activated cell sorting and sequencing system
Researchers at the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences (Qingdao, China) have developed a Raman-activated cell sorting and sequencing (RACS-SEQ) instrument that enables functional identification, sorting, and sequencing of individual cells in a label-free manner.
Fluorescence-activated cell sorting (FACS) is the most commonly used strategy and instrument for single-cell sorting. However, cells need to be fluorescently labeled (on specific DNA, protein, or metabolite biomarkers) prior to flow cytometry. Therefore, cells without known biomarkers, cells that cannot be fluorescently labeled, and cells that are not yet cultured are all beyond the reach of FACS.
The system is based on the Raman concept and the invention of key technologies including Raman-activated Gravity-driven Cell Encapsulation (RAGE) and Raman-activated micro-Droplet cell Sorting (RADS). It consists of four functional modules, including single-cell Raman imaging, interpretation of Ramanome data for phenotype and function, and RACS-SEQ library construction for the sorted single cells.
The RACS-SEQ system is equipped with an intelligent information system, which consists of the Ramanome Lab Information System (RamLIS), Ramanome Explorer (RamEX), and Ramanome Database (RamDB). Users can acquire Raman data for samples rapidly and accurately through RamLIS, process and mine these data online through RamEX, and store all information for future data storage and mining through RamDB. The single cells sorted by RAGE and RADS can then be subjected to single-cell sequencing via single-cell nucleic acid extraction and amplification in microdroplets.
The RACS-SEQ system consists of the instrument and the accessory kits, and supports RACS and the directly coupled single-cell sequencing. (Image by Ji Yuetong)
The system, via novel technologies such as RAGE and RADS, not only retains cell activity post-sorting, but also greatly elevates the quality of single-cell genome assemblies. The genome coverage of a single bacterial cell can reach 95% using the RACS-SEQ system, according to Ma Bo, group leader of Microfluidics Systems at the Single-Cell Center. The released RACS-SEQ system also includes a number of kits such as the single-cell clinical antimicrobial resistance testing kit, the post-RACS single-cell genome amplification kit, and the RAGE microchip kit.
"[This system] will find wide applications in microbiome and synthetic biology research, and support the biomedicine, biosecurity, industrial biotechnology as well as marine biotechnology industries," says Liu Chenli, director of the Center for Synthetic Biology Engineering Research, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences.
The system was released at the 20th Molecular Spectrum Conference of China and the 2018 Annual Conference of Spectroscopy on October 20, 2018.