POINT-OF-CARE MEDICINE/MOLECULAR DIAGNOSTICS: Infectious disease control with portable CMOS-based diagnostics
Infectious diseases constitute a major health threat worldwide: They kill millions of people each year and otherwise affect many more. Such microbial threats include drug-resistant tuberculosis, MRSA, hepatitis, HIV, and Ebola. The toll is especially high in developing countries where aging populations and urbanization compound the challenge.
The broad use of antibiotics, which has become a common practice, further exacerbates the situation. Overuse of antibiotics has shown to be linked to the many cases of surging drug-resistant viral and bacterial pathogens. The research community long ago realized that the world needs a globally affordable method to rapidly and accurately identify infectious diseases. Such a tool would enable physicians to respond quickly and prescribe narrowly targeted antibiotics and anti-viral drugs, which would simultaneously save lives and contain the spread of lethal, drug-resistant pathogens.
The past decade has seen vast advancement in molecular diagnostics technology. Nucleic acid (DNA, RNA) and protein (antibody) tests now allow doctors to get very precise information about the type of virus and bacteria cells behind each case of infectious disease (see image at the top of this page). However, much of the world's population still does not have timely access to these technologies, largely due to the cost and bulkiness of the equipment required. Today's molecular diagnostics systems are designed for use in professional labs. Patient samples collected at a site of care are packaged and transported to these "reference labs" for batch testing—a model that is not well suited for the many small hospitals and clinics in remote areas of developing countries where resources are limited.
Thus, there is a push for portable systems based on the latest molecular methods that allow precise and timely testing of infectious diseases at the point of care. Such a solution should be globally affordable so it can address the needs of developing countries where infectious diseases hit the hardest.
The quest for portable molecular diagnostics
Researchers have tried many methods to achieve small, portable, and automated molecular diagnostics solutions. To this end, much effort has been put into developing novel microfluidics (e.g., DNA microarrays and microfluidic enzyme-linked immunosorbent assay [ELISA] tests) technologies, many of which demand customized chemistry. However, while microfluidics enables reactions between samples and diagnostic assays on a chip, such chips still need to be scanned or "read out" by instruments such as fluorescence microscopes. The small reaction volume and dense reaction sites of microfluidic chips impose on these instruments an even higher performance requirement, making the total solution bulky and expensive.
Most of today's molecular diagnostic chemistry uses optical methods to detect molecular events based on the principle of fluorescence and chemiluminescence signaling. (Electrochemical molecular sensors have shown promise for miniaturization, but they require sophisticated surface chemistry and suffer from stability and specificity problems.) To meet sensitivity requirements, engineers have had to resort to such special-purpose devices as photomultiplier tubes (PMTs) and cooled charge-coupled devices (CCDs), both of which are not only expensive, but also require bulky and sophisticated electronics and cooling systems to support their operation.
But what if the instrumentation, too, was compact?
Ultra low-light CMOS sensor
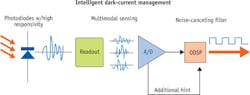
To find a less expensive and smaller alternative, we turn to CMOS image sensors, a low-cost, low-power technology popular for consumer digital and cell phone cameras and capable of unmatched integration. While recent innovations have improved the sensitivity of CMOS image sensors, more improvements in process, circuit, and software are needed to match the sensitivity of PMTs and CCDs. To meet this challenge, engineers at Anitoa Systems (Palo Alto, CA) use novel manufacturing process technology and circuit design, as well as software algorithms that intelligently remove the unwanted noises in CMOS image sensor.
With this approach, Anitoa has fabricated a CMOS image sensor, called ULS24, built on a 0.18 μm CIS technology from a leading specialty foundry (see Fig. 1). The chip has been shown to achieve 3e-6 lux detection sensitivity (at 550nm wavelength), while consuming less than 30mW of power. ULS24 is capable of detecting just a few molecules labeled with fluorescence reporter probes. It forms the basis of a miniaturized quantitative polymerase chain reaction (qPCR) system: The CMOS sensor is paired with LEDs as the optical excitation source to achieve fluorescence-based molecular sensing in a very compact platform.DNA quantification and qPCR
The ULS24 CMOS bio-optical sensor (see Sidebar) has been tested in a variety of applications, including qPCR and DNA detection and quantification-one of the most common procedures in molecular biology. It is frequently performed to evaluate the yields of DNA during purifications and amplification procedures such as PCR, or to detect residual DNA in samples such as drug and protein preparations.
The traditional method for measuring nucleic acid concentration is the determination of absorbance at 260 nm (A260). Major disadvantages of the method are its relative insensitivity (an A260 of 0.1 corresponds to a 5 μg/mL dsDNA solution), and its inability to distinguish between signals from DNA and RNA. Fluorescence nucleic acid stains such as PcioGreen (Life Technologies) fluoresce upon binding to double-stranded DNA. The reagent forms the basis of a highly sensitive DNA assay kit, which has found wide acceptance in research for several reasons including the fact that dye-based fluorescence assays are up to 10,000 times more sensitive than UV absorbance measurements.
Testing and results
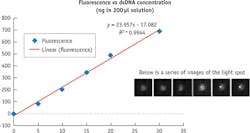
The fluorescence imager includes the items listed in the sidebar "Molecular imager setup." Testing involved first preparing a series of concentration samples containing 0, 5, 10, 15, 20 and 30 nanograms (ng) of double-stranded DNA (dsDNA). These samples are then mixed with Quant-iT Picogreen assay in a series of qPCR tubes. Fluorescent readouts were recorded for each sample and repeated several times; in between, the system was "blanked" using 0 ng sample to create a dark image. We chose a fixed integration time to establish the signal gain of the system, and found 200 ms to be adequate.
When the samples are imaged, each tube containing the sample creates a light spot in the captured image. Captured images were processed by averaging the digital readout from the pixels that define the light spot. The results of analyzing samples are shown in Figure 3. The line describing the concentration dilution series had a correlation coefficient r=.9944. Closer examination of the curve demonstrates that the linear detection range of the PicoGreen assay using the ULS24 extends over three orders of magnitude.Using a SnR of ~10dB—or 3X background noise (standard deviation of measurement results)—we determined that the assay sensitivity for the gain setting of 200mS integration was 1.0 ng. This represents >500 fold improvement in sensitivity over the standard A260 method. (Longer integration time can be chosen to achieve even higher sensitivity.)
The results showed that a CMOS ultra-low light imager combined with wavelength filters provides a very sensitive solution for detecting DNA stained with fluorescence dye. Not only can this method measure the presence and quantify DNA at very low concentration, it does so with a low-power excitation light source such as an LED, and with a short integration time. This not only saves system power and speeds detection, but it also reduces risks associated with photobleaching, a common phenomenon among molecular diagnostic assays that can significantly reduce assay repeatability.
Containing infectious diseases
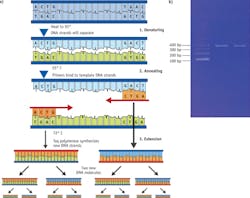
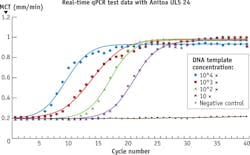
When it comes to detecting very small amount of pathogenic molecules, such as DNA molecules released from viruses or cancerous cells, it is important that the detection method used is not only sensitive but also specific (see Fig. 4).Quantitative PCR achieves sensitivity and specificity through combined amplification and detection; qPCR can cause target DNA strands be selectively replicated millions of times, with the help of an enzyme called polymerase whose activity is modulated by temperature. As the target DNA strands are replicated they bind with specially designed molecular probes labeled with fluorescence materials (see Fig. 5).
The imager leverages microfluidic innovations, and provides many benefits by offering fast integration time, complementary imaging capability, high sensitivity, and good signal-to-noise ratio. With this combination, we are now in the process of building a palm-sized qPCR system that will work with today's off-the-shelf qPCR diagnostic assays.
For the future, we envision small and portable molecular diagnostics devices deployed at point-of-care worldwide, enabling rapid, on-site diagnostics of infectious disease so that doctors can make quick decisions about treatment and prevent further spread of these diseases. Such devices will be Internet-enabled, and the diagnostic results will be transmitted to a central database in the cloud, allowing doctors, drug companies, and policymakers to make better-informed decisions on global epidemic control.
Molecular imager setup
Components of the molecular imaging system include:
• ULS24 CMOS ultra low-light sensor (Anitoa Systems)
• 25-mm-diameter objective lens with 17 mm focal length, <10 degree angle of incidence (AOI)
• Et525/50m emission filter, OD6, with center wavelength 525 nm and 50 nm bandwidth (Chroma Technology)
• Blue (470 nm) Rebel Start LED light source, 70l m at 700 mA, filtered with Et470/40x bandpass filter, OD6 (Chroma Technology). Center wavelength 470 nm and 40 nm bandwidth
• Real-time PCR reaction tube (Eppendorf)
• Quant-iT PicoGreen dsDNA Assay Kit P11496 (Life Technologies)
Zhimin Ding | CEO
Zhimin Ding is the president of Anitoa Systems, LLC (Menlo Park, CA).