Fluorescence analysis method boosts DNA and RNA visualization
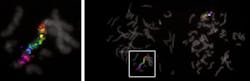
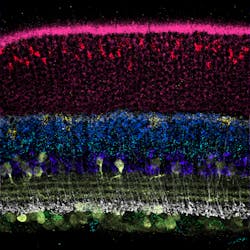
A team of researchers from Harvard University’s Wyss Institute for Biologically Inspired Engineering and Harvard Medical School (HMS; Cambridge, MA) has developed a highly programmable and practical method that significantly enhances the sensitivity, customization, and multiplexing capabilities of fluorescence in situ hybridization (FISH) analysis. The team demonstrated the method, dubbed Signal Amplification by Exchange Reaction (SABER), to amplify FISH signals at distinct RNA and DNA targets in the different layers of the mouse retina, and visualize up to 17 different target regions on the human X chromosome simultaneously. In addition, they used SABER to identify a genomic element that controls the transcription of a specific gene expressed in a particular type of retinal neuron.
Researchers have been using FISH analysis for decades to find specific DNA and RNA sequences in intact cells and tissues within their vast seas of nucleic acid molecules. Because of its ability to light specific sequences up under the microscope at the exact locations at which they reside, FISH has come to be a go-to method in the diagnosis of chromosomal abnormalities, investigation of the 3D organization of genomes in cells' nuclei, and analysis of the immediate products of gene expression known as messenger RNAs.
However, it remains challenging to pick up sequences with FISH that are rare, especially in thick tissues that emit unspecific background fluorescence, and to detect many targets at the same time in multiplexed analysis. Several methods have been developed that can amplify weaker FISH signals, but these cannot be easily customized, are unable to simultaneously visualize a large number of DNA or RNA target molecules, and they are expensive and difficult to use.
"With SABER-amplified FISH analysis and its sensitivity, multiplexing potential, practicality, and cost efficiency, we created a way to overcome key limitations of existing methods and provide researchers with a more powerful handle on a broad spectrum of analyses, ranging from fundamental research, to biomarker discovery and the development of therapeutics," says Wyss Institute Core faculty member Peng Yin, Ph.D., who corresponded the study together with Constance Cepko, Ph.D., the Bullard Professor of Genetics and Neuroscience in the Blavatnik Institute at HMS, and Brian Beliveau, Ph.D., a former Damon Runyon Postdoctoral Fellow in Yin’s group and now Assistant Professor at the University of Washington’s Department of Genome Sciences (Seattle, WA).
Related: 'DNA origami' technology boosts labeling options for fluorescence microscopy
In SABER, the researchers first programmed the Primer Exchange Reaction (PER) method previously reported by Yin’s group to synthesize a longer concatemer of identical shorter sequences with the help of a catalytic self-folding DNA hairpin structure. This engineered mechanism shares similarity with the naturally occurring enzymatic mechanism that extends “telomeres” at the ends of chromosomes with identical DNA repeats to protect them from degradation but can be executed in a test tube.The PER-generated concatemers are then hybridized via short complementary handle sequences to their target DNA or RNA sequences in fixed cells and tissues where they provide a scaffold with multiple binding sites for short fluorescent oligonucleotides (imagers) added next.
"In contrast to conventional FISH analysis, SABER allows us to not only bind one, but many fluorescent dye molecules to a single nucleic acid target, which can considerably boost the strength of the signal coming from its location inside the cell, and we can even further amplify it by creating branched structures in extra hybridization steps, with additional concatemers grown from internal branch points of already existing concatemers," says Jocelyn Kishi, Ph.D., a co-first author on the study and Postdoctoral Fellow working with Yin who developed SABER with Beliveau.
"SABER enables us to tune the signal strength to the abundance and localizations of specific RNA and DNA targets in FISH analysis, and, because concatemers for multiple targets can be synthesized in bulk ahead of time, SABER provides us with standardized probe sets that can be easily generated and re-used in different studies," Beliveau says. "In addition, by repeatedly washing imagers complementary to one set of targets out of a sample and substituting them for imagers that bind to other targets in a process we call DNA-Exchange, we were able to further multiplex our approach."
The spectral features of distinguishable fluorescent dyes allow researchers to only analyze up to four targets at the same time by conventional fluorescence microscopy. With DNA-Exchange, a set of imagers can be washed out of a sample and replaced by a different one binding to PER-generated concatemers at different target sites, and the process can be repeated multiple times. Using the multiplexing potential of ‘Exchange-SABER’, the team visualized 17 distinct regions of the human X chromosome in one fell swoop using the same type of microscopic equipment.Applying SABER to thick tissues, Cepko's group at HMS demonstrated the performance and utility of the method in the retina, which is organized into layers with a high diversity of neural cell types.
"We previously applied single-cell sequencing to dissociated retinal tissue, but were unable to use the resulting markers to identify many cell types simultaneously in intact tissue," says co-first author Sylvain Lapan, Ph.D., a Postdoctoral Fellow in Cepko's group. "By providing multiplexed detection of marker transcripts, SABER is allowing us to close this loop and add a spatial dimension to our analyses."
Key to their analysis was a method developed for the study by co-first author Emma West that, based on cell surface fluorescence stainings, can computationally map all cells contained in a cross-section of the retina to specific positions. This enabled the team to assign individual signals obtained by SABER-enhanced FISH analysis to exact 3D spaces occupied by cells.
"By modeling cellular coordinates and transcript positions, we can create a digital representation of our tissue that is quantitative at the single cell level. This is not only useful for identifying cell types, but also for analyzing phenotypes and the activity of introduced genetic elements," says West, who is a Graduate Student in Cepko's group.
In addition, the team used SABER-FISH as an effective tool to identify genomic elements known as enhancers that drive gene expression in specific retinal cell types. “We introduced recombinant DNA constructs into the retina in which those elements were individually coupled to a reporter, and by performing SABER-FISH analysis to quantify the copy numbers of constructs in single cells, as well as the expression of the reporter, we were able to correlate activity of one specific enhancer with cell type-specific expression," Lapan says.
"The sensitivity and simpler, cheaper workflow of SABER-FISH allow us to gather much more information from experiments in which we manipulate the retina in order to understand the regulatory networks that govern its development, and to assess the efficacy of our gene therapy approaches to eye diseases," Cepko says. "Importantly, this approach has the potential to similarly enhance research in many other tissue types and organs."
Full details of the work appear in the journal Nature Methods.