New imaging technology brings human genome into view
A combination of high-resolution microscopy and advanced computational modelling is capturing some of the most intricate views of the human genome to date. This could offer a much deeper understanding of human genes as well as their impact on health and diseases.
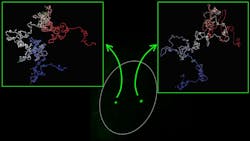
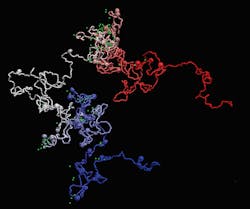
The new technique—developed by researchers at the Centre for Genomic Regulation and the Institute for Research in Biomedicine, both in Barcelona—takes images of the structure of the human genome and shows how individual genes fold at the nucleosome level. This is allowing the researchers to virtually navigate 3D models of genes and see details of their movements and flexibility (see video).
The research began with the development of another imaging technology: immuno-OligoSTORM (iOS); this allows the researchers to simultaneously visualize genes and proteins in super-resolution, says Vicky Neguembor, a staff scientist in the Cosma Lab at the Centre for Genomic Regulation, who is corresponding author of the research published in Nature Structural & Molecular Biology.
This approach makes use of Oligopaints—probes that are fluorescently-labeled, single-stranded DNA oligonucleotides (short, single strands of synthetic DNA or RNA) that can be used to visualize a range of genomic regions—and OligoSTORM—stochastic optical reconstruction microscopy, which is a widely used super-resolution imaging technique based on single-molecule localization—to label the genes of interest.
It also uses DNA point accumulation in nanoscale topology (PAINT), a super-resolution technique for binding and unbinding of DNA imager strands, to label proteins in super-resolution with stochastic optical reconstruction (STORM) microscopy—a widely used super-resolution method based on single-molecule localization.Next, the Barcelona-based team developed a technology called Modelling iOS (MiOS).
“This bridges the information coming from our iOS images with sequencing-based information to model how genes are folded in 3D in the most comprehensive and resolved manner to date,” Neguembor says.
Existing methods can only reach a modelling resolution of about 5 kb. With MiOS, the researchers can model gene structures at single nucleosome resolution, which provides a much deeper understanding of gene folding. And while other techniques combine information from Hi-C (a technique to measure the frequency at which two DNA fragments physically associate in a 3D space, linking chromosomal structure directly to the genomic sequence) and/or imaging of DNA, Neguembor says her team’s method also incorporates MNase-seq (micrococcal nuclease digestion with deep sequencing, which measures nucleosome occupancy in the C. elegans genome) and super-resolution imaging of histones.
“This allows us to determine how nucleosomes are organized along the DNA fiber within a gene of interest,” Neguembor says. “Since nucleosomes are key regulators of gene expression, making genetic regions more or less accessible and understanding how they are distributed within a gene is extremely useful to decode gene function.”
In developing their strategy, the team can also identify changes in conformation occurring in genes that are fundamental for human development.
“By reaching these goals, we can visualize how genes that change their activity during human development also undergo important structural reshaping,” Neguembor says.
The researchers are further developing the MiOS technology, in particular, by adding more functionality to detect how transcription factors—proteins involved in the process of converting or transcribing DNA into RNA, for example—bind to DNA.
In the future, MiOS models could predict what happens to genes when things go wrong; for example, by cataloging variations in the shape of genes that cause disease. The technology could also be used to test drugs that change the shape of an aberrant gene, helping discover new treatments for different types of disease. MiOS could also help decode how genes’ folding is reshaped during physiological processes such as tissue growth or regeneration.
About the Author
Justine Murphy
Multimedia Director, Digital Infrastructure
Justine Murphy is the multimedia director for Endeavor Business Media's Digital Infrastructure Group. She is a multiple award-winning writer and editor with more 20 years of experience in newspaper publishing as well as public relations, marketing, and communications. For nearly 10 years, she has covered all facets of the optics and photonics industry as an editor, writer, web news anchor, and podcast host for an internationally reaching magazine publishing company. Her work has earned accolades from the New England Press Association as well as the SIIA/Jesse H. Neal Awards. She received a B.A. from the Massachusetts College of Liberal Arts.