Recent advances are allowing photomultiplier-tube detectors to meet the challenging requirements of developing biological research applications such as DNA sequencing, which demand instrumentation with highly sophisticated detector technology.
JAMES A. ROBERTS
The sequencing of the human genome was completed fairly recently and already subsequent projects such as the SNP consortium (an international group characterizing single nucleotide polymorphisms [SNPs]-common DNA sequence variations among individuals-to treat human disease; snp.cshl.org) and the related HapMap (www.hapmap.org) are probing deeper and revealing new information. This knowledge may provide an understanding of the intricate molecular interactions occurring within cells, encoded by the genome, and ultimately yield a molecular-level understanding of the difference between health and disease in individuals. Extending this knowledge to drug development, disease diagnostics, and therapeutics may one day revolutionize medical care. The realization of this goal rests in part upon development of instrumentation that provides faster and more specific access to genomic information.
DNA sequencing
Sequencing the genome was to a large extent dependent upon basic methods developed by Nobel Laureate Fred Sanger in the 1970s. Fundamentally, the method consists of the separation, identification, and quantification of amplified, radio-labeled DNA fragments. Today the labels are fluorescent dyes incorporated during the amplification process, with each dye indicating the presence of a DNA fragment terminated with one of the four DNA bases: adenine (A), cytosine (C), guanine (G) or thymine (T). In addition to gaining an understanding of gene expression and function, DNA-sequencing applications include areas such as forensics and the development of techniques for detection of hazardous biological organisms like anthrax.
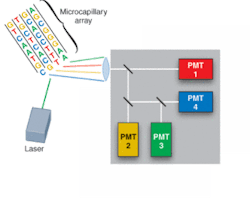
FIGURE 1. Commercial DNA-sequencing instruments use laser-induced fluorescence to identify the adenine (A), cytosine (C), guanine (G), or thymine (T) base of DNA fragments. The fragments are labeled with fluorescent dyes, loaded into a multicapillary electrophoresis array, migrate according to size/charge in the applied electric field, and are identified as they pass an observation point.
Driven by the Human Genome Project, development of DNA-sequencing instrumentation has realized enormous improvements in capability, with high-throughput instruments capable of sequencing several million bases of DNA per day. In a typical commercial DNA-sequencing instrument, DNA fragments labeled with fluorescent dyes are loaded into a multicapillary electrophoresis array and migrate according to size/charge in the applied electric field. As the fragments pass an observation point, laser-induced fluorescence gives rise to an optical signal identifying the A, C, G, or T base (see Fig. 1).
The detection problem
Demands for more information, at faster speeds and lower costs-in short, at higher throughput-have driven many instrument development efforts to extend the basic multicapillary design. Tremendous improvements in instruments have increased the number of capillaries, from the standard 96 to 384 and even 768, and decreased the size of capillaries while reducing the required sample concentration. With these developments, instrument performance hinges on a decreasing concentration of fluorescently labeled DNA migrating more quickly through more and smaller capillaries, andremaining within the excitation region for a shorter duration. These constraints significantly reduce the amount of fluorescence signal available and place increased demands on the detection system. Hence, a detection system for DNA-sequencing instruments must satisfy the demand for high sensitivity and speed under inherently low-light conditions.
The detection solution
Photomultiplier tubes (PMTs) have been widely used in commercially available DNA-sequencing instrumentation. Their performance characteristics-low dark noise, noise-free gain, excellent low-light sensitivity, and fast response time-make them ideally suited for such applications.
Fundamentally, PMTs are vacuum-tube devices with performance resulting from the basic structure consisting of a photocathode, a series of dynodes, and an anode. In a PMT, incident photons strike the photocathode to generate photoelectrons, which are amplified via the dynode chain and ultimately arrive at the anode, producing an output signal.
The spectral responsivity of a PMT is dependent upon the window material, but more significantly for DNA-sequencing applications, the visible responsivity is determined by the photocathode. A fundamental measure of the sensitivity of the photocathode to incident light is determined by the quantum efficiency (QE)-the ratio of photoelectrons produced relative to the number of photons incident at the photocathode. The other determinant of performance for PMTs is the gain. Gain in the PMT depends upon the application of voltage between the anode and photocathode and results from the sequential amplification of the initial photoelectron by the dynodes. This process gives rise to amplification of the signal by a factor of 106.It is the gain that gives a PMT excellent low-light sensitivity.
An assessment of the use of PMTs on the basis of performance for typical DNA-sequencing conditions merits a look at the sources of noise in PMTs and a basic evaluation of their signal-to-noise ratio (S/N). In a PMT there are two primary sources of noise. The first, the dark-current noise, is the noise associated with photoelectrons created in the absence of any light signal and is the critical factor in determining the lower limit of light detection. The other source is the shot noise of the signal, which arises from the statistical variation in the incident light signal. Other contributions are noise from the external amplification electronics and the secondary emission process.
Because DNA-sequencing measurements are not conducted in a photon-limited regime and because the gain increases the signal above any external electronic amplification noise contributions, the sole noise consideration is the shot noise of the signal. As a result, a basic expression for a PMT signal-to-noise ratio can be given as:
S/N = IAS/(2qIASFB)1/2
In this expression, IAS is signal at the anode, q is the charge of the electron, F is the excess noise associated with the gain, and B is the observation bandwidth. The S/N will therefore increase with an increase in IAS or a decrease in B. Increasing IAS can result from an increased light intensity via more signal photons or increased photocathode quantum efficiency. It is important to note that the gain is dependent upon the applied voltage and that the gain increases the signal level; however, it is not a contributing factor in the S/N because it contributes equally to the signal and to the noise.
High gain is necessary for detection as the signal level becomes extremely low. In this limit, better signal to noise can be achieved with photon counting. For this configuration, discrimination of signal from noise is accomplished by the use of a high-speed amplifier and a discriminator. The output-current signals of the PMT are converted to voltage by the amplifier and the discriminator then rejects low-amplitude pulses. In this way, the high gain of the PMT gives rise to signal pulses with higher amplitude, allowing for easier setting of the discrimination level to optimize the signal to noise.
Improving the solution
To address the signal limitations posed by increased throughput demands, new developments in PMT engineering have produced designs that offer significant performance enhancement. Enhanced spectral response is available through the incorporation of new semiconductor photocathode materials like gallium arsenide (GaAs) and gallium arsenide phosphide (GaAsP). In fact, GaAsP detectors have quantum efficiency approaching 50% in the visible range (550 nm), while GaAs photocathodes have very appreciable quantum efficiency of about 10% at 800 nm. These photocathode developments, in combination with the use of a longer wavelength or fluorescence-resonance energy-transfer (FRET) tags-chemically attached markers that aid in the detection of a certain molecule-can give rise to significant signal enhancement, reducing the drawbacks of limited fluorophore concentration.
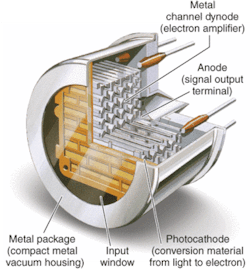
FIGURE 2. The metal-channel dynode design of metal-package PMTs allows multichannel outputs that can simultaneously detect fluorescence from multiple molecules, increasing detection sensitivity and speed.
Metal-package PMTs, another recent development, incorporate a metal housing in place of the traditional glass housing of PMTs (see Fig. 2). While the metal-channel dynode design of these PMTs offers high speed, the greatest advantage of the design for DNA sequencing (in addition to significant miniaturization) is the multichannel output capability resulting from multiple anodes within a single device. The increase in performance by the simultaneous detection of the fluorescence from each of the four dyes reduces the need for complicated optical paths, which can dramatically impact not only signal intensity but also speed.
Another feature arising from the small size of metal-package PMTs is the development of compact assemblies with the electronic circuitry for power and voltage dividing as well as signal processing built into a single package. Modules having integrated application-specific integrated circuits (ASICs) for special functions such as multichannel array amplification and discrimination for photon counting, dramatically reduce complexity and size, and allow straightforward incorporation of PMTs within existing instrument designs.
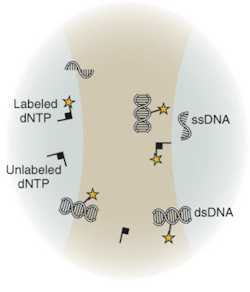
Underscoring the tremendous advances that have already been made with existing techniques and instrumentation, the National Human Genome Research Institute at the National Institutes of Health (NIH; Bethesda, MD) has spearheaded an aggressive initiative toward development of completely new technologies that will drive down costs of sequencing, with the future goal of a $1000 genome. Technologies under development differ from the traditional capillary array techniques. Sequence-by-synthesis techniques already show promise for dramatic improvements in throughput and reduction in cost. Many research groups are actively developing techniques that may not only reduce cost but also revolutionize DNA sequencing. Several of these techniques are based on the observation and/or sequencing of DNA at a single-molecule level. Others address detection of DNA target sequences by fluorescence correlation spectroscopy (FCS; see Fig. 3) or evanescent-field fluorescent techniques for observation of dye-labeled DNA bases.
These new techniques promise enormous improvements to DNA sequencing through the potential for highly parallel platforms. And, unlike more conventional technologies, single-molecule techniques are not limited by read-length (the minimum number of molecules in a string that can be detected) or electrophoretic separation hurdles. In principle, future instrumentation based on single-molecule detection offers the potential of examining gene expression within single cells. At the very low-light levels encountered in these single-molecule techniques, photon-counting PMTs are ideally suited for making the measurements.❏
JAMES A. ROBERTS is responsible for university sales and marketing at Hamamatsu, 360 Foothill Rd., Bridgewater, NJ 08807; e-mail: [email protected]; www.sales.hamamatsu.com