Machine-intelligence algorithms analyze images
Machine-intelligence algorithms analyze images
James O. Gouge and Sally B. Gouge
Image metamorphosis is a method for handling images with a special set of machine-intelligence-based algorithms rather than an orthodox mathematics-based approach. The technique can be used to analyze photographic, computer-graphic, and medical-tissue images. The technique is most useful, however, with the type of abstract images constructed from nonvisual data produced by backscatter detection such as ultrasound, radar, and sonar.
With conventional mathematics-based methods, including standard edge/contrast enhancement, filtering algorithms, convolutions, histograms, and fast-Fourier-transform-based approaches, images are dealt with as arrays of numbers. These arrays are blindly processed to yield subsequent arrays that have no meaning until they are presented to the human visual cortex as picture elements, or pixels, to produce perceived images. This approach does not take into account the visual relationship of these elements to each other in an image. While mathematics-based rote methodology yields some meaningful results with many types of images, it is less successful for dealing with complex, abstract, and color images.
Perception
Consider how the human brain addresses images: the human visual cortex deals with large groups of pixels and their relationships to each other to form an Ointelligently perceivedO image. Better understanding of complex images can thus be gained by evaluating them with machine-intelligence-based methods similar to human processes. To further convey the greatest amount of information, the resulting images should also be presented in concert with these perception processes.
Because machine-intelligence-based algorithms may change the form and content of the resultant processed images, we have dubbed the technique Oimage metamorphosisO algorithms to set them apart from the conventional image-processing approach. In the basic analysis of a pixel group, the algorithm evaluates a target pixel and its near and far neighboring pixels along one of eight radials. Each of the pixels along the radial selected has a numerical value (gray scale) of backscatter luminance. The radial-pixel information forms determinants that describe the target pixel and are processed individually or in concert, depending on the desired application or function.
The radials are used to construct reproducible and recognizable patterns of information. The shape-extraction algorithm searches each radial for an edge, or luminance change. Edge positions relative to the target pixel are stored as individual bit strings. Processing the eight radials creates an accumulated string of bits that correlates to the radial shape of the edges. These edges can be looked at as dots that are connected to form a shape. A radial contour-extraction algorithm measures changes along the edges identified by the shape-extraction algorithm.
A separate algorithm measures the asymmetry between opposing radial pairs extending from the target pixel. The gray-scale value differences are accumulated, producing a number that correlates with the overall symmetry of the pixels surrounding the target as well as indicating the asymmetry of the feature being imaged.
On the edge
The metamorphosis approach for analyzing radial-pixel-luminance variations from that of the target pixel provides a straightforward method for complex image processing. The edge-data bit strings require relatively little processing time once translated to the computer. For the image as a whole, edges, lines, and contours are extrapolated and compared to a backscatter-pattern data base, a process that can, for example, emphasize distinctions between critical areas of cells or tissue differences.
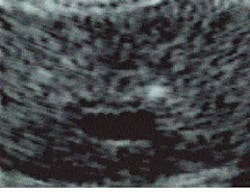
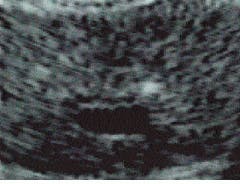
BioComputer Research (Summerville, GA) is using image metamorphosis in a number of research projects dealing with anomaly detection and changes in ultrasound images of various tissues (see images on p. 146). In these applications, the algorithm-processed results are further refined by neural-network techniques in which the images are compared, via a fuzzy-logic data base, to actual images of biopsied tissue. The pattern data base itself is thus further expanded.
A major clinical advantage of metamorphosing images is that the highlighted tissue becomes easier to target with a biopsy needle. A physician can examine areas of interest that normally would be difficult or impossible to identify amid the gray overcast of the average ultrasound image. Images can also be made immediately after surgery to ensure completeness of cancerous-tissue removal, even in the presence of severe tissue surgical trauma.
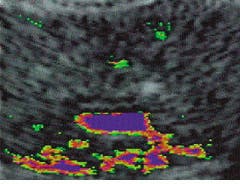
To aid the physician in obtaining a Odirect hitO with his biopsy needle, we have combined intelligent algorithms with a proprietary digital-signal-processor (DSP) coprocessor board and a commercially available frame grabber. The DSP runs in real time, thus producing metamorphosed images during the entire ultrasound examination. Sam Graham, chief of urology at Emory Clinic (Atlanta, GA) has used the system in a recent study to evaluate images of normal and anomalous tissue in the human prostate gland. He notes that image metamorphosis is a promising Oalternative method to the subjective visual analysis of transrectal ultrasound imaging.O
Unprocessed axial transrectal ultrasound image of a prostate gland (left) is metamorphosed to highlight anomalous/abnormal tissue (right). The tissue is differentiated, highlighted, and quantified at the single pixel level?from the smallest anomaly (green) to the largest (red)?indicating the amount of tissue echoic deviation from normal. Blue indicates urine, blood, and low-density fluids. The two small foci of early-stage cancer (green) seen at the upper right and left sides of the prostate, as well as other features, are not visible or distinguishable in the raw image.