PHOTONIC FRONTIERS: GENOMICS: Advances in photonics-based tools expand genomics possibilities
Genomics—the mapping, analysis and study of genomes—has been on a roll since the successful sequencing of the human genome a decade ago. Rapid improvements in technology have slashed sequencing costs and improved data quality. The National Human Genome Research Institute (Bethesda, MD) says the cost of sequencing a person's complete genome dropped from $95 million in September 2001 to $7700 in October 2011. A new generation of instruments should reduce the cost to $1000 in a few years, according to a National Research Council study released in August, "Optics and Photonics: Essential Technologies for Our Nation" (also known as "Harnessing Light 2.0").1
The driving force behind the heavy investment in genomics is the hope that understanding the genome could bring a revolution in medical diagnosis and treatment. But genomic research is already paying off in basic science, answering key questions about evolutionary relationships, and revealing that a few percent of the genes in modern Europeans come from Neanderthals.
Photonics has helped make this progress possible. In the 1970s, DNA sequencing was a painfully slow process that could identify fewer than 1000 base pairs a day by labeling DNA fragments with radioactive phosphorus and separating them with gel electrophoresis. At that rate, sequencing the whole 3-billion-base-pair human genome would have taken some 8000 years. Replacing radioactive tags with fluorescent ones and improving separation methods multiplied capacity a factor of 10 in the 1980s. Further advances have slashed costs while speeding sequencing to a couple of billion base pairs a day. And new laser techniques are emerging for manipulating genetic material and synthesizing genes, chromosomes, or whole genotypes of living organisms from the raw building blocks of DNA.
DNA sequencing basics
DNA is a double helix, formed by two chains of nucleotides coiled around each other. It encodes genes as a sequence of "bases" that are attached to the backbone chain and that also form a hydrogen bond with a second base on the other backbone chain. The base adenine (usually denoted as A) bonds to thymine (T), and cytosine (C) bonds to guanine (G). This selective bonding makes the two strands of DNA complementary, so decoding one strand identifies the sequence of the second strand. The base sequences encode genes, which perform biological functions in the organism but also include other stretches of DNA with no known function.
Sequencing DNA requires a way to identify which bases are attached to the chain. Initially this was done by tagging bases with radioisotopes, but tagging the bases with dyes that fluoresce at different wavelengths made the process much simpler. Sensors could record the colors produced by illuminating the DNA with a laser or other light source, making it possible to read out the sequence of bases present.
The optical part of the process is relatively simple; much of the work is in developing chemical processes to analyze the DNA base-by-base. Typically, chromosomes are first split into two strands then chopped into random-length pieces containing hundreds to thousands of bases. Those fragments then are sequenced in large volume, and computers match overlapping DNA sequences to stitch the pieces together and sequence an entire gene or chromosome.
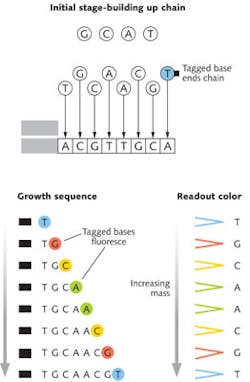
Details differ widely among techniques, but a good example of how sequencing works is the chain-termination approach that Frederick Sanger of the University of Cambridge (Cambridge, UK) developed in the 1970s. Many copies of segments of a single strand of DNA are mixed with the A, C, G, and T bases needed to assemble a complementary strand to produce a double helix. The mixture also includes a special version of one base modified, so it terminates the chain by preventing the addition of more bases and carrying a tag to identify it.
This produces a variety of DNA chains of different lengths, each tagged with a dye identifying the final base. These are separated by mass then illuminated to produce their characteristic fluorescence and read out the base sequence, as shown in Fig. 1. Changing the mass-separation stage from gel electrophoresis to polymer gels in capillary tubes in the early 1990s greatly increased speeds for the Human Genome Project, as described in a special report in Laser Focus World's May 2001 issue.2 With further improvements, this venerable process now can read nearly 1000 base pairs with accuracy to 99.999%, and sequencing costs as low as $0.50 per thousand bases.3
Since completion of the genome project, a new generation of sequencing techniques has achieved higher sequencing rates using different chemistry and massive parallelism, albeit for shorter DNA chains. Some rely on laser illumination of fluorescent tags to identify bases; others rely on energy transfer from adenosine triphosphate (ATP) to luciferase to produce light.4
MALDI and mass spectrometry
The new sequencing techniques are designed for large-scale projects, but they can be too costly for quickly sequencing short lengths of DNA containing only a few genes. The older Sanger technique offers an alternative, but it has problems reading some sequences. To overcome those limits, Chunmei Qiu and colleagues of Columbia University (New York, NY) have adapted a technique called matrix-assisted laser desorption/ionization (MALDI), which had been used for mass spectrometry of proteins.
MALDI uses an ultraviolet (UV) or infrared (IR) laser to produce a hot plume containing delicate biomolecules and to ionize them for time-of-flight mass spectrometry, which can sequence DNA chains not amenable to the Sanger process. So far they have demonstrated sequencing of chains with up to 37 bases.5
Optical tweezers and DNA manipulation
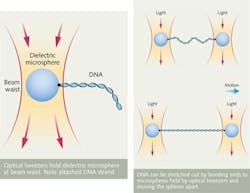
Laser light also can manipulate biomolecules using optical tweezers. As shown in Fig. 2, a laser beam, tightly focused by a lens or microscope objective with numerical aperture higher than 1.2, concentrates light to a high intensity at the beam waist, producing a high electric-field gradient, which attracts dielectric objects to the center of the beam. The effect works best for biomaterials at near-IR (NIR) wavelengths where they are transparent, producing a balance of forces that holds the object near the beam center. Moving the beam carries the object along with the beam.Bare DNA molecules are small compared to the wavelength of light, making them hard to hold and manipulate, but they can be attached to larger-sized beads, also shown in Fig. 2. This allows studies of the forces applied to DNA during transcription and other cellular processes.6 Last year researchers at the University of Michigan (Ann Arbor, MI) reported using constant-force optical tweezers to study the effects of sequence variations on the mechanical properties of DNA.7 And a group at the University of Cambridge (Cambridge, UK) used optical tweezers to study DNA molecules inside a nanocapillary.8
Synthetic biology and laser printing of DNA
So far laser tweezers remain laboratory instruments, but the provocative claims of "DNA laser printing" by a stealthy startup called Cambrian Genomics (San Francisco, CA) raise interesting questions about optical manipulation to assemble DNA.
It's part of a hot and controversial field called "synthetic biology," in which molecular biologists build DNA molecules from short sequences or individual bases, either to insert into existing organisms or to create new organisms.9 So far the most visible success in the field has been the J. Craig Venter Institute's (Rockville, MD) creation of what it called "the first synthetic cell" two years ago, based on copying the genome of a natural bacteria.10 But the process was very slow, error-prone, and very expensive.
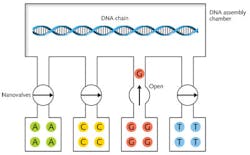
Cambrian Genomics is trying to do better, and reduce error rates to near zero by resequencing the DNA after the initial synthesis.11 CEO Austen Heinz told me in a brief phone interview that the company could print sequence-verified strings of 100 base pairs. He would not disclose details, but said "you could figure [them] out for yourself."A search of Heinz's past publications provides a few clues. Before founding the company, he worked on optofluidic systems with Sunghoon Kwon at Seoul National University (Seoul, South Korea). Heinz coauthored a paper on the use of UV light to control flow through optofluidic valves.12 That technique might be useful in DNA synthesis, as shown in Fig. 3, but it's not laser printing. Indeed, because base pairs measure only about 0.3 nm it's hard to envision how light could manipulate them individually.
Time will tell if Cambrian Genomics can deliver on its promises, but it's clear the field as a whole is well worth watching.
REFERENCES
1. G. Overton, "SPIE and OSA applaud National Academy Optics and Photonics report (Harnessing Light 2)," Laser Focus World online (Aug. 14, 2012); http://bit.ly/MX38cI.
2. V. Coffey, "DNA sequencing provides the key to the map of man" in "Special Report: Exploring the Genome," Laser Focus World, 37, 5, 137–145 (May 2001); http://bit.ly/S7kQ8Q.
3. J. Shendure1 and H. Ji, "Next-generation DNA sequencing," NatureBiotechnol., 26, 1135 (Oct. 2008); doi:10.1038/nbt1486.
4. M. Kircher and J. Kelso, "High-throughput DNA sequencing - concepts and limitations," Bioessays, 32, 524–536 (2010); doi:10.1002/bies.200900181.
5. C. Qiu et al., "Design and synthesis of cleavable biotinylated dideoxynucleotides for DNA sequencing by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry," Analyt. Biochem., 427, 193–201 (2012); doi:10.1016/j.ab.2012.04.021.
6. D.J. Stevenson, F. Gunn-Moore, and K. Dholakia, "Light forces the pace: Optical manipulation for biophotonics," J. Biomed. Opt., 15, 041503 (July/August 2010).
7. K. Raghunathan et al., "Mechanics of DNA: Sequence Dependent Elasticity," Proc. SPIE, 8097, 80970C (2011); doi:10.1117/12.895297.
8. O. Otto, L.J. Steinbock, and U.F. Keyser, "Nanocapillaries and Optical Tweezers for Studies on DNA in Confinement," Proc. SPIE, 8097, 80970B (2011); doi: 10.1117/12.894703.
9. Synthetic Biology web site, http://synthetic biology.org.
10. D. Gibson et al., "Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome," Science, 329, 52–56 (2010); doi:10.1126/science.1190719.
11. S. Kotler, "Holy Genetically-Engineered Organisms Batman - Synthetic Biology Has A Banner Month!" Forbes online (July 17, 2012); http://onforb.es/PsUr7I.
12. S.H. Lee et al., "Active Guidance of 3D Microstructures," Small, 6, 2668–2672 (2010); doi:10.1002/smll.201001248.
About the Author
Jeff Hecht
Contributing Editor
Jeff Hecht is a regular contributing editor to Laser Focus World and has been covering the laser industry for 35 years. A prolific book author, Jeff's published works include “Understanding Fiber Optics,” “Understanding Lasers,” “The Laser Guidebook,” and “Beam Weapons: The Next Arms Race.” He also has written books on the histories of lasers and fiber optics, including “City of Light: The Story of Fiber Optics,” and “Beam: The Race to Make the Laser.” Find out more at jeffhecht.com.